The main objective of the present study was to isolate phytohormone-producing, phosphate-solubilizing strains of Azospirillum from wheat to be used as inoculants for plant growth promotion. Five Azospirillum strains were isolated from the rhizosphere of field-grown wheat (Triticum aestivum L.), and it was confirmed by BOX-polymerase chain reaction (PCR) that the isolates were different and not re-isolates of the same strain. Sequence analysis of the PCR-amplified 16S rRNA gene indicated that four isolates showed maximum similarity to Azospirillum brasilense and one isolate showed maximum similarity to Azospirillum zeae. This is the first report indicating the presence of an A. zeae like isolate in the wheat rhizosphere in Pakistan. The bacterial isolates were characterized for their plant growth-promoting traits, phosphate solubilization, and indole-3-acetic acid (IAA) production. None of the isolates showed phosphate solubilization activity in the commonly used Pikovskaya medium. However, all strains (except AzoK4) exhibited ability to solubilize tricalcium phosphate (TCP) in modified Pikovskaya medium in which sucrose was replaced by Na-malate, as well as in TCP-supplemented Luria-Bertani (LB) medium. Organic acids, such as acetic, citric, lactic, malic, and succinic acids, were detected in culture supernatants of the tested Azospirillum strains. All strains exhibited ability to produce IAA in the growth medium, except Azospirillum sp. AzoK1. Among the strains tested, the maximum IAA production (30.49±1.04mgL−1) and phosphate solubilization (105.50±4.93mgL−1) were shown by a pure culture of Azospirillum sp. AzoK2. In pot experiments, single-strain inocula of Azospirillum sp. AzoK1 and AzoK2 improved wheat plant growth.
Plant growth-promoting rhizobacteria (PGPR) isolated from the rhizosphere or root surfaces comprise an active component of biofertilizers. Several direct or indirect mechanisms are adopted by these microorganisms to enhance the plant growth, including biological nitrogen fixation, solubilization of insoluble minerals, production of phytohormones, plant relief from stress, and protection against pathogens.1 Among PGPR, strains of Azospirillum are most extensively studied for their beneficial effects on the growth and yield of many agronomically important crops.2 It has been reported that inoculation with these diazotrophic bacteria reduces the use of chemical nitrogen fertilizers.3 Indole-3-acetic acid (IAA) production by Azospirillum spp. has been proposed to play a major role in root proliferation and consequent plant growth promotion.4 IAA is produced by Azospirillum during all growth stages, and its synthesis continues well after the stationary phase.5 This feature makes the bacteria especially qualified for plant growth promotion when the effect lasts weeks or months after inoculation. The amount of IAA produced varies greatly among different species and is also influenced by culture conditions, the growth stage, and availability of a substrate(s).6 It was reported that wheat inoculation with Azospirillum strains resulted in a yield increase of 31%.7 Inoculation with Azospirillum also increases the uptake of mineral nutrients such as N, P, K, and Ca by wheat.8
Soils usually contain sufficient reserves of phosphorus to support plant growth if P is in available forms. Unfortunately, phosphorus may be present in water-insoluble forms when it is not readily accessible to plant roots. Therefore, there is a dire necessity to make phosphorus more available to plants.9 Phosphorus solubilization activity has been reported for many genera of PGPR, including Azospirillum. PGPR produce organic acids, which convert insoluble phosphorus into available forms, i.e., primary and secondary orthophosphates.10,11 The organic acids produced by PGPR include malic, acetic, citric, oxalic, lactic, formic, gluconic, and 2-keto-gluconic acids.12,13 In pure bacterial cultures, organic acid production has been demonstrated upon utilization of various sugars and sugar alcohols, such as sucrose, glucose, fructose, d-xylose, and mannitol.10 Phosphate-solubilizing microbes are considered important members of PGPR, and their application as biofertilizers has been shown to improve growth of cereals and other crops.13
In the present study, five Azospirillum strains were isolated from the rhizosphere of wheat, followed by strain identification using DNA-based techniques [BOX-polymerase chain reaction (PCR) and analysis of 16S rRNA gene sequences]. After confirmation of IAA production, P solubilization and amplification of the nifH gene, as effects of inoculation with selected Azospirillum strains was studied on wheat plants.
Materials and methodsStudy siteThis study on isolation of plant growth-promoting azospirilla continued for 16 months (from January 2012 to April 2013) at the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan. The NIBGE is located in a subtropical area of Pakistan (31°23′36.2″N and 73°01′39.9″E) at an elevation of 184m.
Isolation of Azospirillum strains from the rhizosphere of wheatIsolation of Azospirillum-like strains from the rhizosphere of wheat (Triticum aestivum L.) was carried out by an enrichment culture technique using semi-solid, N-free malate (NFM) medium.14 Twenty-five rhizospheric soil samples were collected from field-grown plants at the tillering stage, i.e., 40 days after sowing. Soil suspensions were prepared by adding 1g of soil to 10mL of water, and 100μL was transferred to 1.5mL microcentrifuge tubes containing 0.9mL of semi-solid NFM medium. The inoculated microtubes of NFM were incubated for 24–72h at 30°C. The growth of Azospirillum-like cells showing typical spiral motility was detected under a light microscope at 1000× magnification (Akon, Japan). Six consecutive transfers from inoculated to fresh NFM microtubes were made to obtain an enriched culture. Serial dilutions (10×) of the enriched culture were made, and 100-μL aliquots of 10−3–10−5 dilutions were spread on NFM agar plates and incubated for 4–6 days at 30°C. Bacterial colonies were picked from the plates, transferred to NFM semi-solid medium, and incubated for 24–48h at 30°C. Bacterial growth obtained in NFM medium was streaked on Luria-Bertani (LB) agar plates,15 and the plates were incubated at 30°C for 24–72h to obtain single colonies. After repeated streaking and transfer of single bacterial colonies, five pure isolates were obtained. For long-term storage, bacterial growth obtained in LB broth was kept in the medium with 15–20% glycerol at −80°C.
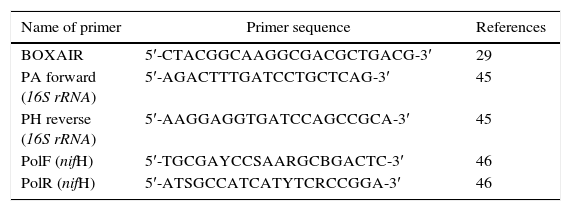
Differentiation of Azospirillum strains by BOX-PCRAll chemicals, except the primers used for PCR, were purchased from Fermentas, Germany. Each PCR reaction mixture (25μL) contained 0.15μL of 5UμL−1Taq DNA polymerase, 5μL of 10× Taq polymerase buffer, 5μL of 25mM MgCl2, 2μL of 5mM deoxynucleotide triphosphates (dNTPs), 1.5μL of 10μM BOXAIR primer (Table 1), and 25ng of DNA template extracted by CTAB method.16 The reaction mixture was incubated in a thermocycler (Eppendorf, Germany) at 94°C for 5min, followed by 30 cycles of 94°C for 1min, 42°C for 1min, and 72°C for 1.5min, and final extension at 72°C for 10min. Amplified PCR products of different sizes were separated on 1% agarose gel electrophoresis and stained with ethidium bromide. Gels were viewed under ultraviolet (UV) light and photographed using a gel documentation system (CN-1000/26MX, Vilber Lourmat, France).
Amplification of 16S rRNA and nifH genesFor amplification of the 16S rRNA and nifH genes, a reaction mixture of 25μL, containing 15ng of template DNA, 0.12μL of 5UμL−1Taq DNA polymerase, 2.5μL of 10× Taq polymerase buffer, 1μL of 5mM dNTPs, 2μL of 25mM MgCl2, and 1μL of each 10μM forward and reverse primers, was prepared. The primer set PH/PA was used for amplification of 16S rDNA, and the primer set PolF/PolR was used for amplification of nifH (Table 1). The program used for PCR amplification of 16S rDNA included following steps: initial denaturation at 94°C for 5min, followed by 30 cycles at 94°C for 1min, 54°C for 1min, and 72°C for 2min, and final extension for 10min at 72°C. The same thermal conditions were used for amplification of nifH, except that the annealing temperature was 48°C.
Identification of isolates by 16S rRNA sequence analysisThe PCR products of the 16S rRNA gene were eluted from an agarose gel using the Gene JET™ Extraction Kit (Fermentas, Germany), and the purified PCR products were sequenced commercially by Macrogen, South Korea. The sequence data were assembled and analyzed using the Lasergene sequence analysis software package (DNA Star, Inc., Madison, WI, USA). Sequence similarity searches using the Basic Local Alignment Search Tool (BLAST) were performed by comparing the sequences obtained to other microbial sequences in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/BLAST/). For construction of a phylogenetic tree, the 16S rRNA gene sequences of type strains and other strains closely related to our five isolates were obtained from the NCBI database. Multiple sequence alignments were performed by the Clustal_X program.17 Phylogenetic trees were constructed by the maximum-likelihood method based on the Jukes–Cantor model18 using MEGA version 6.19 All positions containing gaps and missing data were eliminated. There were a total of 816-base pair (bp) positions in the final dataset.
Quantification of IAA produced by Azospirillum strainsFor detection and quantification of IAA produced by the bacterial isolates, cultures were grown in Erlenmeyer flasks containing 40mL of LB medium supplemented with l-tryptophan (100mgL−1) at 30°C for 5 days. The supernatants of the cultures were obtained by centrifugation of stationary-phase cultures at 12,000×g for 15min, and the alkaline (pH 8.3) supernatants were adjusted to pH 2.8 with 1N HCl. Auxins were extracted from the acidified culture medium with equal volumes of ethyl acetate,20 evaporated to dryness, and re-suspended in 1.5mL of methanol. The samples were passed through 0.2μm filters (Orange Scientific GyroDisc CA-PC, Belgium) and analyzed by high-performance liquid chromatography (HPLC, Varian Prostar), with the system equipped with a UV detector and a C-18 column. A mixture of methanol:acetic acid:water (30:1:70, v/v/v) was used as a mobile phase at a rate of 0.6mLmin−1.21 Pure IAA was used as a standard. The IAA in the samples was identified and quantified by comparing the retention time and peak area with those of the standard using computer software (Varian Chromatography Systems, Walnut Creek, CA, USA).
Quantification of soluble phosphorus and organic acids in the growth mediumFor the estimation of soluble phosphorus, bacterial cultures were grown in three different media, i.e., Pikovskaya medium,22 modified Pikovskaya medium, and LB medium supplemented with tricalcium phosphate (TCP, 5gL−1). In modified Pikovskaya medium, sucrose was replaced with sodium malate (10gL−1), which is a preferred C-source for azospirilla.14 The bacterial cultures were grown in conical flasks containing 40mL of the media at 30°C for 7 days. Cell-free supernatants were obtained by centrifugation of stationary-phase cultures at 12,000×g for 15min. The supernatants were analyzed to estimate soluble phosphorus by the molybdate blue color method.23
Organic acids were extracted from cell-free culture media by mixing the supernatant with an equal volume of ethyl acetate. The ethyl acetate phase was separated and evaporated to dryness. Organic acids were re-suspended in 1.5mL of methanol and filtered through 0.2μm filters (Orange Scientific GyroDisc CA-PC, Belgium). The samples were analyzed on a Perkin Elmer (PE) series 200 HPLC system with 20μL auto-sampler PE NELSON 900 series interface, a PE NELSON 600 series link, and PE NCI 900 Network Chromatography interface, followed by detection using a diode-array detector at 210nm and UV spectra (190–400nm). A Cation H Micro-Guard pre-column, followed by an Aminex HPX-87H analytical column, were used for separation. Sulfuric acid (0.002molL−1) was used as a mobile phase with a flow rate of 0.6mLmin−1. Citric acid, malic acid, acetic acid, succinic acid, gluconic acid, lactic acid, and oxalic acid (100ppm solutions) were used as standards. The peak areas and retention times of the samples were compared with those of the standards for quantification of organic acids produced by the bacterial strains.
Inoculation of wheat plants with Azospirillum strainsThree Azospirillum strains, AzoK1, AzoK2, and WB3, were used as inoculants for wheat plants (variety Sehar-2006) grown in non-sterile soil pots (0.02m3). The pots were kept in a net house under natural light and temperature conditions. Azospirillum strain WB3 (previously isolated from the rhizosphere of wheat)24; was used as a positive control. The pots were filled with 10kg of soil (about 27cm deep) of the Hafizabad series (Aridisol-coarse-loamy, mixed, hyperthermic Ustalfic, Haplargid in the US Department of Agriculture classification). The soil was collected from the top 30cm of an experimental field of the NIBGE. The pH of this clay loam was 8.3, total N was 0.61gkg−1, organic matter was 6.2gkg−1, and available P was 68mgkg−1. Ten healthy seeds of equal size were sown in five different places at equal distances in each pot. Each treatment was replicated five times. A nitrogen fertilizer at 80% of the recommended dose (1.2g of urea/pot) was applied to the uninoculated control as well as to all inoculated treatments in two equal split doses, i.e., at the tillering (30 days after sowing) and flowering (75 days after sowing) stages. Before sowing, 80% of the recommended dose of P (0.6g of diammonium phosphate/pot) was applied to the soil. Bacterial cultures for plant inoculation were grown in 100mL conical flasks containing 40mL of LB medium at 30°C for 24h. Cell pellets were obtained by centrifugation at 12,000×g for 10min, and the pellets were re-suspended in 40mL of sterilized water for use as inocula. One milliliter of inoculum (1×108cells) was added to each plant after 1 week of germination, and 1mL of sterilized water was applied to the uninoculated controls. The plants were harvested at maturity (180 days after sowing). Plant data (dry weight and grain weight) for 10 plants from each treatment were measured after drying the plant material at 65°C for 2 days.
Estimation of phosphorus and nitrogen contents in plant materialFor estimation of nitrogen contents in shoots and grains, a method described by Van Schouwenberg and Walinge25 was followed. Measurement of phosphorus contents in shoots and grains was carried out by the ammonium vanadate–molybdate method.26
Statistical analysisEffects of the bacterial isolates on growth parameters of wheat were determined by a completely randomized design analysis of variance (ANOVA) using the Statistix 8.1 software. Mean values and standard errors were also calculated. The means were compared by Fisher's least significant difference test at α=0.05 for all parameters.
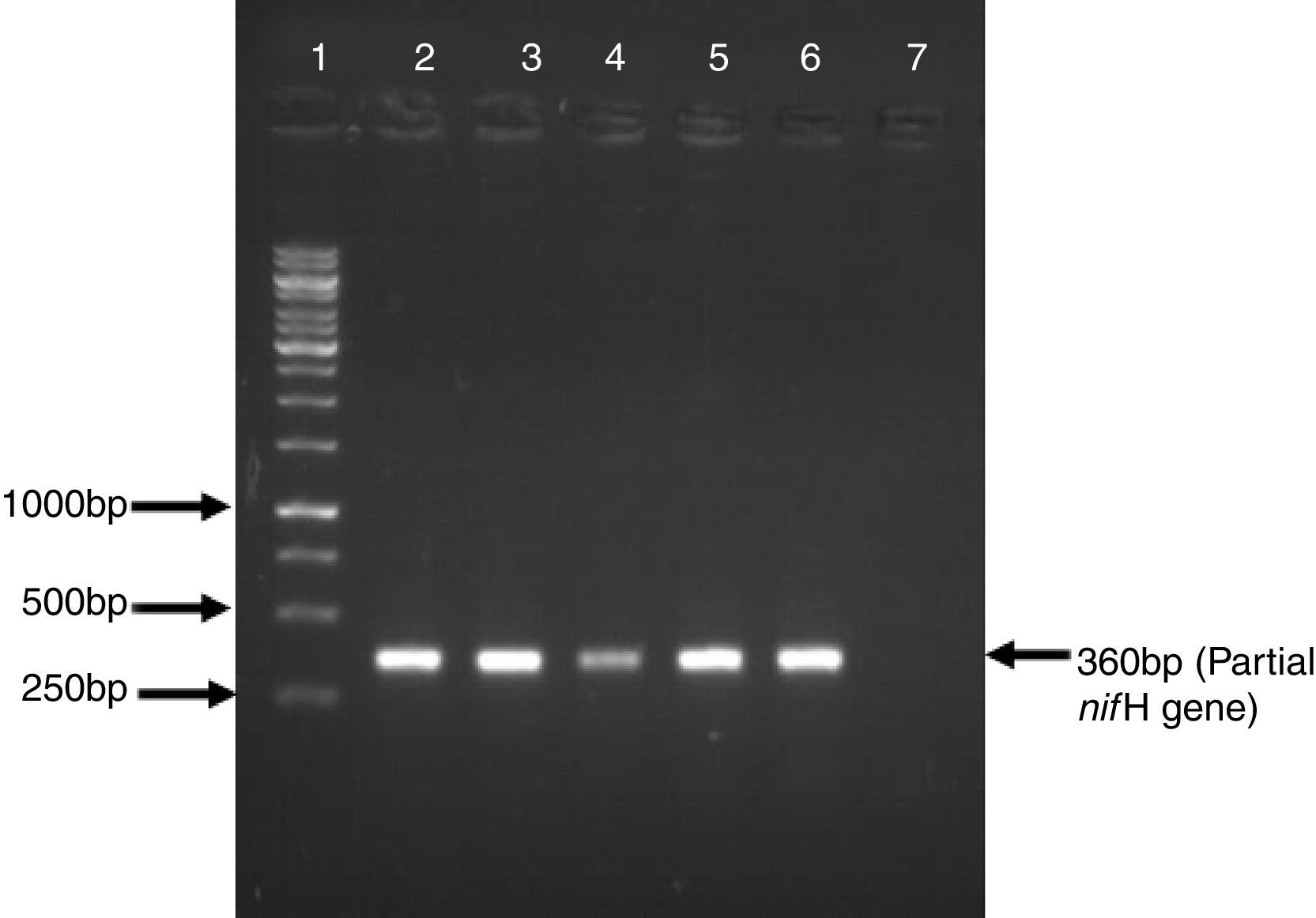
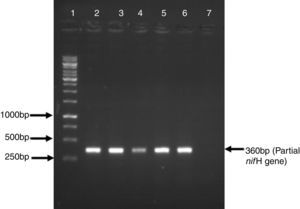
ResultsFive Azospirillum-like strains were isolated from the rhizosphere of wheat. All the isolates formed pinkish colonies on LB agar plates. The cells were rod-shaped and showed spiral motility typical of azospirilla. To further strengthen this observation based on morphological characteristics, a partial sequence of the nitrogenase-encoding gene nifH was amplified, which is present in all nitrogen fixers, including azospirilla. A DNA band of the expected size (360bp) was observed in all strains (Fig. 1).
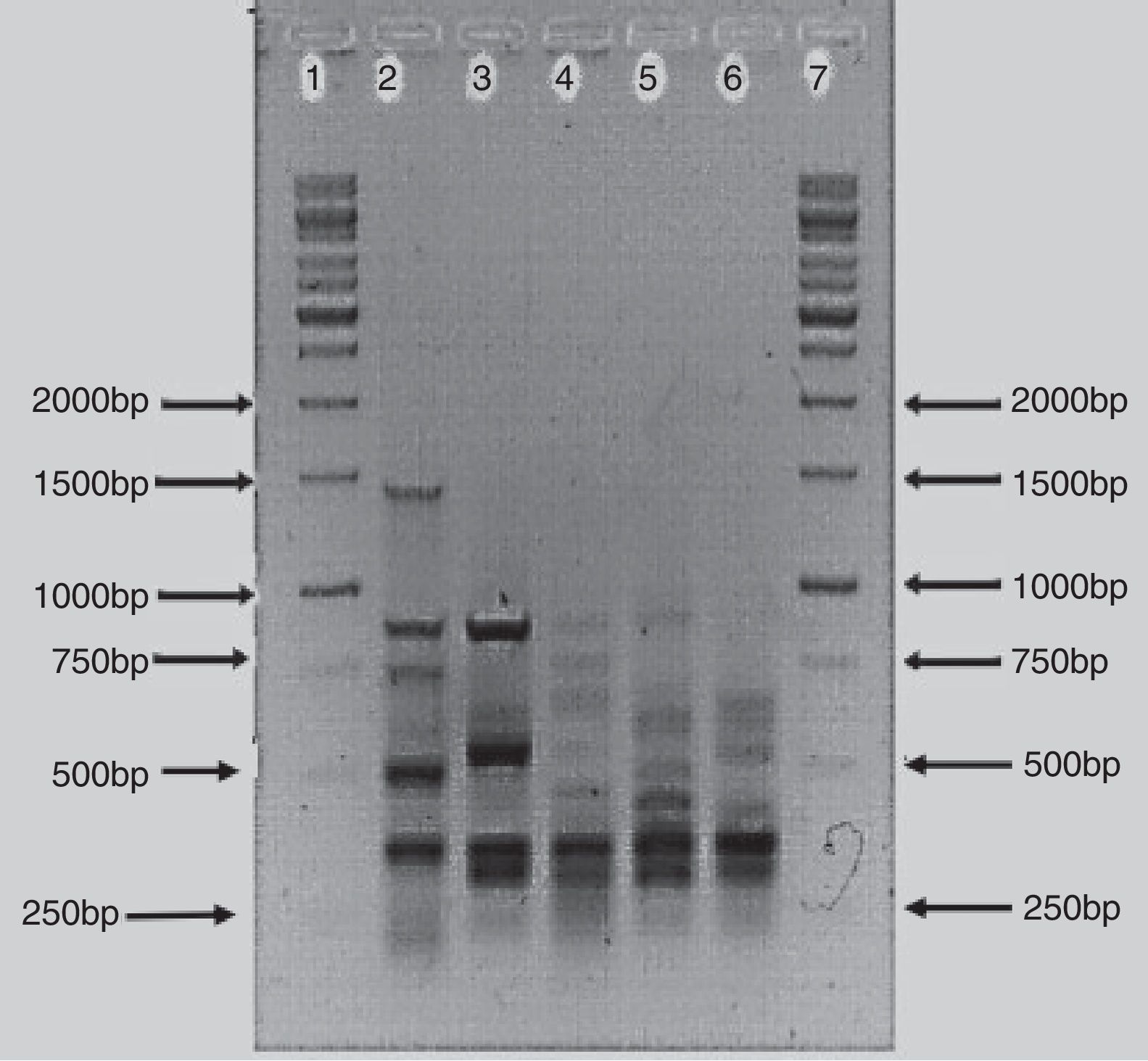
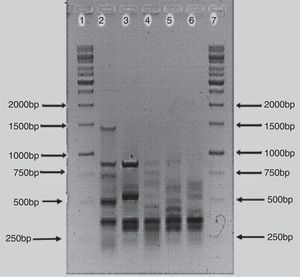
The BOX-PCR analysis confirmed that the bacterial isolates were different strains and not re-isolates of the same strain. All five strains showed unique banding patterns of the PCR products (Fig. 2). The banding pattern of isolate AzoK1 showed that it was totally different from the other strains. The sizes of the bands were between 250 and 1500bp. One strong DNA band (approximately 300bp) was present in four strains but absent from AzoK1. Many minor bands of different sizes were also amplified from the four strains.
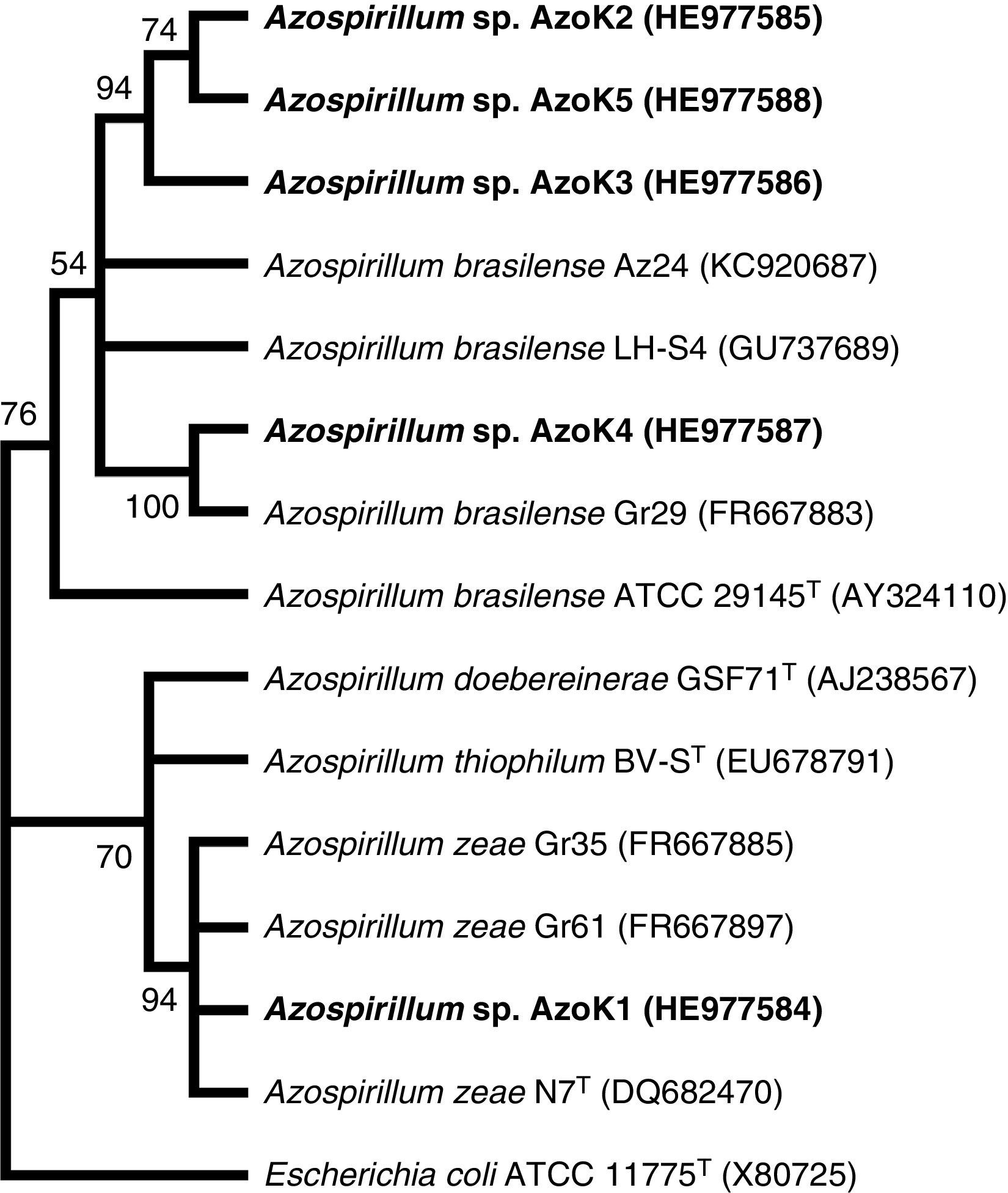
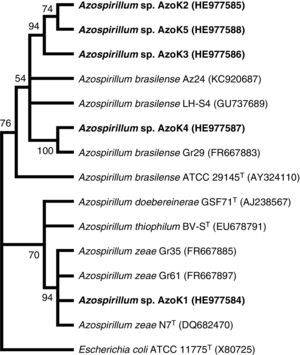
Azospirillum strain AzoK1 showed maximum 16S rRNA sequence homology (99%) to Azospirillum zeae, and the other four isolates shared higher sequence homology with Azospirillum brasilense strains. The constructed phylogenetic tree based on the 16S rRNA gene sequences showed that isolate AzoK1 shared a cluster with A. zeae (Fig. 3). Three isolates, AzoK2, AzoK3, and AzoK5, formed a cluster with A. brasilense strains, and isolate AzoK4 fell in a separate cluster within the main group of A. brasilense strains.
Phylogenetic tree based on 16S rRNA gene sequences. The tree was constructed by maximum likelihood method. Bootstrap values over 50% (based on 100 replications) are shown at each node. Accession numbers are given in parentheses. The isolates obtained in the present study are given in bold letters.
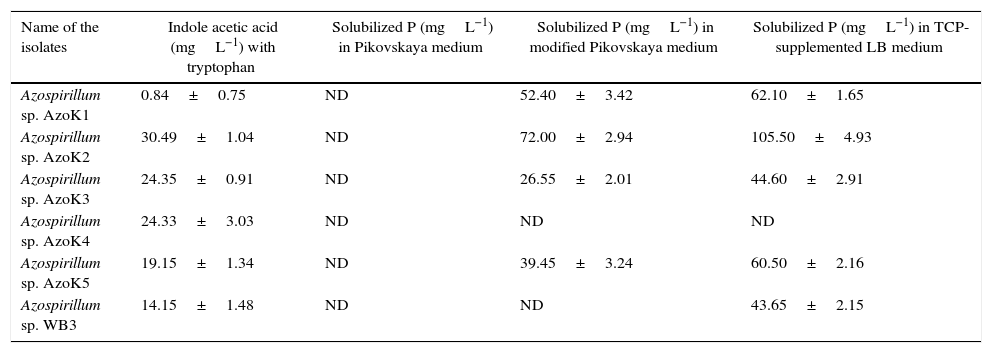
IAA production was estimated in LB broth supplemented with l-tryptophan (Table 2). The maximum amount of IAA was produced by Azospirillum sp. AzoK2, followed by strains AzoK3 and AzoK4. Only a minor amount of IAA was detected in the culture of Azospirillum sp. AzoK1. The quantity of phosphate solubilized by the bacterial strains was determined in three different growth media, i.e., Pikovskaya medium, modified Pikovskaya medium, and TCP-supplemented LB medium. In Pikovskaya medium, phosphate solubilization was not detected for any strain after 7 days of incubation (Table 2). However, phosphate-solubilizing activity of the isolates was detected in modified Pikovskaya medium and TCP-supplemented LB. In modified Pikovskaya medium, the maximum amount of phosphate was solubilized by Azospirillum sp. AzoK2. Phosphate solubilization was not detected in modified Pikovskaya medium for Azospirillum sp. WB3 and AzoK4. All the Azospirillum strains solubilized phosphate in TCP-supplemented LB, except Azospirillum sp. AzoK4 (Table 2). The maximum amount of phosphate was solubilized by Azospirillum sp. AzoK2.
Phosphate solubilization and indole acetic acid (IAA) production by Azospirillum strains (Mean ±SD).
| Name of the isolates | Indole acetic acid (mgL−1) with tryptophan | Solubilized P (mgL−1) in Pikovskaya medium | Solubilized P (mgL−1) in modified Pikovskaya medium | Solubilized P (mgL−1) in TCP-supplemented LB medium |
|---|---|---|---|---|
| Azospirillum sp. AzoK1 | 0.84±0.75 | ND | 52.40±3.42 | 62.10±1.65 |
| Azospirillum sp. AzoK2 | 30.49±1.04 | ND | 72.00±2.94 | 105.50±4.93 |
| Azospirillum sp. AzoK3 | 24.35±0.91 | ND | 26.55±2.01 | 44.60±2.91 |
| Azospirillum sp. AzoK4 | 24.33±3.03 | ND | ND | ND |
| Azospirillum sp. AzoK5 | 19.15±1.34 | ND | 39.45±3.24 | 60.50±2.16 |
| Azospirillum sp. WB3 | 14.15±1.48 | ND | ND | 43.65±2.15 |
ND, not detected; SD, standard deviation.
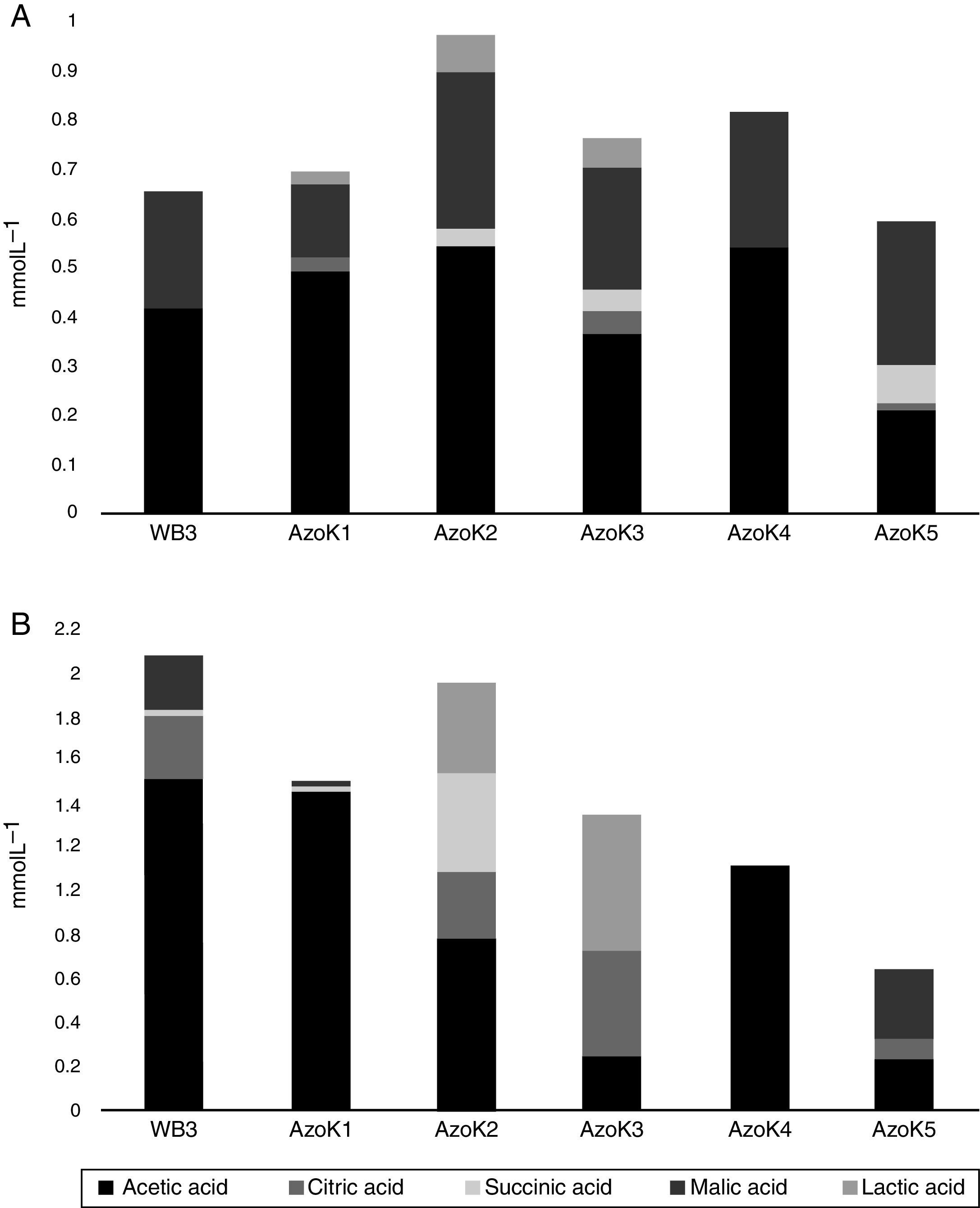
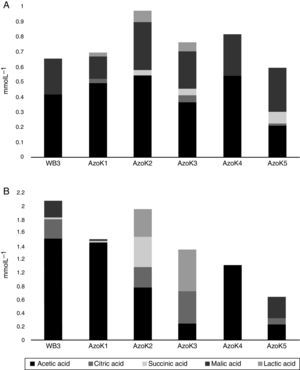
Organic acids were detected in cell-free supernatants of modified Pikovskaya medium and TCP-supplemented LB after 7 days of incubation. Acetic, citric, lactic, malic, and succinic acids were detected in almost all Azospirillum cultures. In modified Pikovskaya medium, acetic and malic acids were detected in higher amounts in the cell-free supernatants of all Azospirillum strains, but relatively minor amounts of lactic acid were present in this medium (Fig. 4A). In TCP-supplemented LB medium, acetic acid was produced by all strains in significant amounts (Fig. 4B). The maximum amounts of citric acid were produced by three isolates, except AzoK1 and AzoK4, in this medium. A significant amount of succinic acid was only produced by AzoK2, and malic acid was one of the major acids detected in the supernatants of two strains, WB3 and AzoK5.
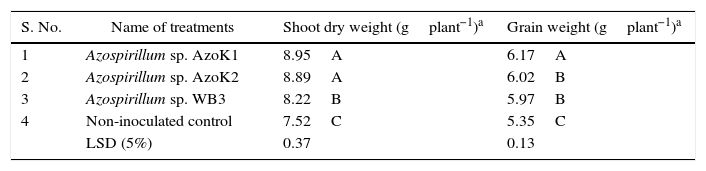
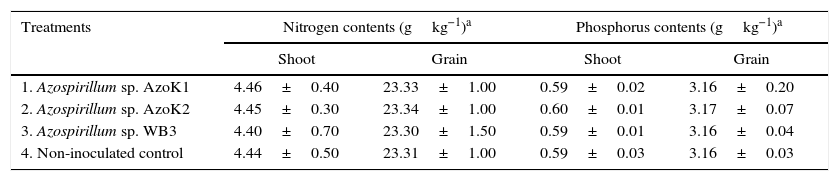
The maximum plant shoot dry weight (8.95±0.20g) and grain weight (6.17±0.11g) were recorded in the strain AzoK1-inoculated treatment, and the increases in the plant shoot and grain weights over the uninoculated control were 19 and 15%, respectively (Table 3). The nitrogen and phosphorus contents in the grains and shoots of the inoculated plants were not significantly different from those of the control plants (Table 4).
Effect of bacterial inoculation on growth of wheat plants in the soil pot experiment.
| S. No. | Name of treatments | Shoot dry weight (gplant−1)a | Grain weight (gplant−1)a |
|---|---|---|---|
| 1 | Azospirillum sp. AzoK1 | 8.95A | 6.17A |
| 2 | Azospirillum sp. AzoK2 | 8.89A | 6.02B |
| 3 | Azospirillum sp. WB3 | 8.22B | 5.97B |
| 4 | Non-inoculated control | 7.52C | 5.35C |
| LSD (5%) | 0.37 | 0.13 |
LSD, least significant difference.
Effect of Azospirillum inoculation on the nitrogen and phosphorus contents of wheat plants (shoot and grain) in the soil pot experiment (mean±SD).
| Treatments | Nitrogen contents (gkg−1)a | Phosphorus contents (gkg−1)a | ||
|---|---|---|---|---|
| Shoot | Grain | Shoot | Grain | |
| 1. Azospirillum sp. AzoK1 | 4.46±0.40 | 23.33±1.00 | 0.59±0.02 | 3.16±0.20 |
| 2. Azospirillum sp. AzoK2 | 4.45±0.30 | 23.34±1.00 | 0.60±0.01 | 3.17±0.07 |
| 3. Azospirillum sp. WB3 | 4.40±0.70 | 23.30±1.50 | 0.59±0.01 | 3.16±0.04 |
| 4. Non-inoculated control | 4.44±0.50 | 23.31±1.00 | 0.59±0.03 | 3.16±0.03 |
SD, standard deviation.
In the present study, the rhizosphere soil of field-grown wheat plants was collected and used for isolation of Azospirillum-like strains, which are known to improve the growth and productivity of many agricultural crop species.27 Azospirilla have been isolated from roots of numerous wild and cultivated grasses, cereals, and legumes and from tropical, subtropical, and temperate soils.28 In the N-free growth medium, which is selective for Azospirillum,14 all the isolates showed excellent growth and characteristic spiral motility. On LB agar plates, they formed pinkish, wrinkled colonies. Furthermore, a partial nifH sequence was successfully amplified from all the isolates, indicating that they are potential nitrogen fixers, similar to azospirilla. The cell morphology and colony color of all the Azospirillum-like isolates obtained in the present study were similar.
BOX-PCR polymorphism patterns have been effectively used for differentiation of bacterial strains.29 All the Azospirillum isolates included in this study exhibited unique, strain-specific banding patterns obtained by BOX-PCR. Azospirillum strain AzoK1 showed a BOX-PCR banding pattern significantly different from those of the other strains. One DNA band of about 300bp was amplified from four strains but was absent from the Azospirillum strain AzoK1 profile, indicating that the latter strain belongs to a different group or species.
On the basis of the sequence analysis of the 16S rRNA gene, isolates AzoK2, AzoK3, AzoK4, and AzoK5 were 96–99% similar to A. brasilense strains, and the fifth strain, AzoK1, showed high sequence homology (99%) with A. zeae. Isolate AzoK1 shared a cluster with the A. zeae type strain, and the other four isolates formed a cluster with A. brasilense strains.
Our findings revealed that the wheat rhizosphere is colonized by A. brasilense- as well as A. zeae-related isolates. This is the first report indicating the presence of an A. zeae-like isolate in the wheat rhizosphere in Pakistan. The A. zeae type strain was originally isolated from roots of maize in Canada.30 Originally, this isolate was identified and reported as Azospirillum lipoferum31 but was later re-identified on the basis of a polyphasic taxonomic approach, including morphological characterization, Biolog analysis, DNA–DNA hybridization, and 16S rRNA, cpn60, and nifH sequence analysis.30 Venieraki et al.32 isolated three Azospirillum strains from the rhizosphere of wheat grown in Greece and showed that the isolates recovered were A. zeae by phylogenetic analysis based on 16S rRNA gene sequences.
All the Azospirillum strains tested in the present study failed to grow and solubilize P in the commonly used Pikovskaya medium. It has been reported that azospirilla cannot utilize disaccharides, such as sucrose, which is added to Pikovskaya medium as a sole C-source.33,34 As a result, the Azospirillum strains did not solubilize hardly soluble phosphorus, such as TCP. Azospirilla use organic acids, such as malic, succinic, a-ketoglutaric, gluconic, and lactic acids, as their preferred carbon sources.33 Therefore, Pikovskaya medium was modified by replacing sucrose with Na-malate and used to study P solubilization by the Azospirillum strains. The tested Azospirillum strains produced organic acids in the growth medium. Acetic acid was produced by all strains, including AzoK4, but the latter showed no P-solubilizing activity. A very small amount of malic acid was detected in all culture supernatants compared to the control, showing that most of the malate added to the modified Pikovskaya medium was consumed by the bacteria during incubation.
Phosphate-solubilizing activity of the Azospirillum strains was also studied in TCP-supplemented LB medium. All the strains solubilized phosphate, except Azospirillum sp. AzoK4. The amount of soluble phosphate, determined in the TCP-supplemented LB medium, was higher compared to the modified Pikovskaya medium. Azospirillum sp. AzoK2 produced the maximum amount of soluble phosphorus, followed by Azospirillum sp. AzoK1. Azospirillum sp. AzoK4 failed to show phosphate solubilization and produced only acetic acid in the TCP-supplemented LB medium. In this growth medium, organic acids, e.g., acetic, citric, malic, lactic, and succinic acids, were produced by the other strains tested in the present study, but gluconic and α-ketoglutaric acids were not detected. Goebel and Krieg35 showed that gluconic acid was not formed during growth of A. lipoferum and A. brasilense strains on fructose and was only detected in an A. lipoferum culture during growth on glucose. In the same study, it was reported that A. brasilense showed only slight uptake of glucose and extremely slow growth on this carbon source. Rodriguez et al.10 reported that Azospirillum strains could produce gluconic acid in vitro when grown on fructose amended with glucose as an inducer of gluconic acid production and also showed in vitro phosphate-solubilizing capabilities. This has suggested that phosphate solubilization by these strains is mediated by glucose or gluconic acid metabolism. Since solubilization of phosphate preceded the detection of gluconic acid in the medium, it is possible that even low levels of the acid (below the HPLC detection limit) started to dissolve sparingly soluble phosphate. Alternatively, consumption of gluconic acid by growing cells could have taken place. Furthermore, it has been reported that Azospirillum halopraeferens, a bacterium that does not use glucose and consequently does not produce acid, can solubilize insoluble inorganic phosphate in vitro by an unknown mechanism.36
Bacterial isolates obtained from the rhizosphere of various plants have been shown to produce IAA in pure culture.37 In the present study, all the bacterial strains produced significant amounts of IAA in the culture medium, except Azospirillum sp. AzoK1. Under natural conditions, plant roots excrete organic compounds, including l-tryptophan, which can be utilized by rhizobacteria for IAA biosynthesis.6 Perrig et al.38 reported IAA production ranging from 2.9 to 10.8mgL−1 by two agronomically important Azospirillum strains. Fatima et al.39 also detected IAA in the range from 19.4 to 30.2mgL−1 in A. brasilense cultures. Venieraki et al.40 isolated two A. zeae strains from the rhizosphere of wheat and found that both produced IAA (29.8 and 194.8mgL−1, respectively). In the present study, the amount of IAA produced by Azospirillum sp. AzoK1 in tryptophan-amended LB was 0.84mgL−1, which is very low compared to previous reports. The higher levels of IAA production reported by Venieraki et al.40 could be due to a different IAA detection method (spectrophotometry rather than HPLC) or to the fact that a different growth medium (NFb) was used for bacterial growth. Azospirillum is a rhizospheric bacterium that has been reported to increase the yield of wheat plants by producing phytohormones and growth regulators and by providing salt stress relief.7 Based on the studies on maize and wheat, Hungria et al.41 proposed that the use of inoculants containing Azospirillum might help reach the goal of reducing the use of chemical fertilizers globally. In the present study, all the inoculated strains showed a positive effect on plant growth compared to the uninoculated control. The maximum increases in shoot dry weight (19%) and grain weight (15%) over the uninoculated control were recorded for the plants inoculated with Azospirillum sp. AzoK1. The nitrogen and phosphorus contents of the grains and shoots of the inoculated plants were not significantly different from those of the control plants, suggesting that the growth promotion might have been mainly due to phytohormone production by the inoculated strains. Prinsen et al.42 reported that inoculation of wheat with Azospirillum resulted in an increase of seed yield by 7.4–10.31% in field trials where no N fertilization had been applied. Díaz-Zorita and Fernández-Canigia43 demonstrated that a liquid formulation of A. brasilense increased the number of harvested grains by 6.1% and the grain yield of wheat by 260kgha−1 (8.0%). Inoculation with Azospirillum has been found to affect early plant and root development, shoot and root dry weight, grain yield, and the nitrogen uptake efficiency of plants.44
In the present study, all the isolates exhibited plant growth-promoting traits in pure culture and also improved plant growth in pot experiments. Therefore, these strains qualify as potential candidates to be used for production of biofertilizers in Pakistan and should be further evaluated as inoculants in field trials.
Conflicts of interestThe authors declare no conflicts of interest.