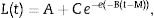Despite the increasing reports on the incidence of fresh vegetables and fruits as a possible vehicle for human pathogens, there is currently limited knowledge on the growth potential of Escherichia coli O157:H7 on different plant substrates. This study analyzed the selective adhesion and growth of E. coli O157:H7 on chili habanero (Capsicum chinense L.), cucumber (Cucumis sativus), radish (Raphanus sativus), tomato (Lycopersicon esculentum), beet (Beta vulgaris subsp. vulgaris), and onion (Allium cepa L.) under laboratory conditions. The Gompertz parameters were used to determine the growth kinetics. Scanning electron microscopy was used to visualize the adhesion of E. coli O157:H7 on the epicarp of the samples. Predictive models were constructed to compare the growth of E. coli O157:H7 on the samples with different intrinsic factors and to demonstrate the low selectivity of the pathogen. No significant difference was observed in the lag-phase duration (LPD), generation time (GT), and exponential growth rate (EGR) of the pathogen adhered to the samples. The interaction between the microorganism and the substrate was less supportive to the growth of E. coli O157:H7 for onion, whereas for tomato and cucumber, the time for the microorganism to attain the maximum growth rate (M) was significantly longer than that recorded for other samples.
Escherichia coli O157:H7 is a foodborne zoonotic pathogen that serving as the causative agent of hemorrhagic colitis and hemolytic uremic syndrome (HUS) in humans. This microorganism possesses a remarkable capability to get transmitted from animal reservoirs (cattle, goats, and sheep) to humans through a variety of food vehicles that plays a critical role in its pathogenicity. E. coli O157:H7 employs multiple mechanisms for colonizing vegetables through adhesion to cell surfaces. The microorganism adheres strongly to tomato skin, spinach leaves and alfalfa sprout roots by curli,1,2 which is mediated by the filamentous type III secretion system composed of EspA filaments.3 Flagella also play a key role in the leaf attachment process in E. coli O157:H7.4 The internalization of E. coli O157:H7 has been demonstrated under controlled environmental conditions in the seedlings of temperate vegetable crops such as lettuce, tomato, cress, radish, alfalfa, and mung bean,5–8 and the adhesion and internalization of E. coli O157:H7 in mature leafy vegetables have been demonstrated both experimentally and in field conditions.9–13
Despite the increasing incidence of fresh produce as a vehicle for human pathogens, there is a dearth of information on the growth potential of E. coli O157:H7 on different plant substrates. Although most of the outbreaks involving the occurrence of E. coli O157:H7 on leafy vegetables have been associated with cross-contamination14–17 there are very few studies that have investigated the adhesion and growth of E. coli O157:H7 in other mature vegetables.18–22 Interestingly, many of these vegetables possess a variety of intrinsic factors that can considerably reduce the growth of foodborne microorganisms; however, E. coli O157:H7 manages to successfully survive under these varying conditions.
Assuming that the degree of mutual interaction between bacterial strain and the specific type of plant affects their association, this study aimed to investigate the capacity of E. coli O157:H7 to colonize freshly consumed vegetables and fruits under experimental conditions. The growth of the pathogen on different substrates was compared using the Gompertz equation to obtain bacterial growth curves and linear regression-derived parameters such as the lag-phase duration, maximum microbial population, and specific microbial growth.
Materials and methodsMicroorganism and preparation of inoculumsEscherichia coli O157:H7 strain (SS11) used in this study was isolated from the feces of pigs from Santa Elena in Yucatán, Mexico.23 The strain was isolated after pre-enrichment with EC broth (Difco, Detroit, MI, USA) using the immunomagnetic separation procedure (IMS) employing anti-E. coli O157 Dynabeads (Dynal Biotech, Norway) as described by the manufacturer. The IMS-enriched bacterial culture (50μL) was streaked onto Chromocult coliform agar (Merck, Darmstadt, Germany) and Sorbitol MacConkey agar (CM813; Oxoid Ltd, USA) supplemented with cefixime-potassium tellurite (SR172; Oxoid Ltd.). The phenotype of the microorganism was characterized with the API 20E system (BioMérieux, France). Strain serotype O157:H7 was identified with the immunocapture system and latex agglutination (Tecra Diagnostics, NSW, Australia). Polymerase chain reaction (PCR) was used to confirm the presence of rfbO157 and stx1 genes in the strain SS11 and to confirm the absence of stx2 verotoxin gene. For this purpose, three pairs of oligonucleotide primers were synthesized for the amplification of rfbO157 (420bp), stx2 (584bp) and stx1 (306bp).24–26 The PCR products were detected by 2% agarose gel-electrophoresis at 100V and staining with 10μg/mL ethidium bromide.
The suspensions of E. coli (104CFU/mL) were prepared for the inoculation of the vegetables and fruits. A single colony of E. coli O157:H7 from a streak plate was suspended in 20mL of EC selective enrichment broth (Difco) and incubated overnight at 37°C (16–18h) in a shaking incubator at 150rpm (Thermo Scientific, USA). The overnight culture (1mL) was inoculated into 99mL of EC broth and incubated at 37°C till it attained the exponential phase (105CFU/mL). The bacterial cell concentrations were estimated by comparing the transmittance of the cell suspension with a previously determined standard curve at 540nm using a UV–visible spectrophotometer. The final concentration of 104CFU/mL was obtained by diluting the bacterial suspension with 400mL of EC broth, and the concentration of the inoculum was further confirmed by plating 0.1mL of an appropriately diluted culture on TSA.
Selection and preparation of vegetables and fruitsThe prepared E. coli O157:H7 suspension was inoculated in triplicate on three vegetables viz., radish (Raphanus sativus), beet (Beta vulgaris subsp. vulgaris), and onion (Allium cepa L.), and on three fruits viz. ripe red tomato (Lycopersicon esculentum L.), cucumber (Cucumis sativus), and green habanero chili (Capsicum chinense L.). Fresh and healthy-looking fruit and vegetable samples were purchased no more than one day prior to the inoculation from a local supermarket and stored at 10°C till use. The samples with visible defects on the surface such as bruises, cuts, or abrasions were excluded from the experiment. The vegetables were decontaminated by immersing in 200mg/L of chlorine solution (6.5mg/L of free chlorine) for 2min before inoculation. The decontamination procedures were previously standardized based on the absence of growth of Enterobacteriaceae on MacConkey agar. The samples were then rinsed with sterile deionized water for 1min and air dried with paper towels in a biosafety cabinet (Esco, Infinity® Class II, Australia).
Growth of E. coli O157:H7 on the epicarp layer of fruits and vegetablesThe samples were treated for five different periods (0, 3, 5, 7 and 9h) of incubation to determine the adhesion capacity of the E. coli O157:H7. The samples were completely submerged into 400mL of E. coli O157:H7 broth (104CFU/mL) for 30min and gently shaken at 100rpm at 37°C. Then the samples were kept individually in sterile air bubble polyethylene bags (Stomacher® Bag, Seward, UK) for the defined period of time for each treatment. After incubation, the samples were washed thrice with sterile deionized water and air dried in a biosafety cabinet. In addition, two control samples were considered. The first one was an internal control of inoculated samples that were not subjected to the final washes, and second one was a negative control of inoculated samples that were decontaminated with 200mg/L of chlorine solution in the final wash.
The adhesion of E. coli O157:H7 to the epicarp of vegetables and fruits was determined by spreading the samples on MacConkey agar. Three slices of the vegetable shell (10g) were cut off aseptically and soaked in 90mL of phosphate buffered saline (PBS, pH 7.1). After a five-fold serial dilution, 0.1mL of the sample solution was inoculated on MacConkey agar in duplicate and incubated for 24h at 35±1°C. The number of colonies observed in the plates was used to calculate the CFU/g of tissue.
Proximate analysisAOAC (Association of Analytical Communities) official methods27 were used for analysis of moisture (Method 925.09), total protein (Method 950.48), fat (Method 983.23), and ash contents of the samples (Method 930.05). pH values were measured by placing the flat electrode (Sensorex, Stanton, USA) directly against the surface of the exposed tissue at three different points. Proximal analysis of the epicarp was performed using the skin of the fruits and vegetables.
Scanning electron microscopy (SEM)The adhesion of E. coli O157:H7 to the exocarp of the samples was visualized using a scanning electron microscope (Philips model XL 30 SEM microscope) operating at an accelerating voltage of 15kV. A final pressure of 4Torr (533.3Pa) was gradually generated in the chamber in order to prevent the samples from drying, prior to performing the analysis.28
Modeling of microbial growthAll the experiments were performed in triplicate. The microbial counts per gram of the sample were converted to log10 values before the calculations. The mathematical modeling of the microbial growth (predicted data) was performed by employing the modified Gompertz equation:
where L(t) is the log of the number of bacteria at time ‘t’ (in hour). A is the asymptotic log of number of bacteria as ‘t’ decreases indefinitely; C is the asymptotic amount of growth (log number) that occurs as ‘t’ increases indefinitely; M is the time (in hour) at which the absolute growth rate is at maximum; and B is the relative growth at time ‘M’.29,30The equation was fitted using a nonlinear regression routine of the SYSTAT statistical computer system package (SYSTAT Inc., Evanston, IL, USA). The algorithm was selected from the set of parameters with the lowest residual sum of squares with a 95% confidence interval. The Gompertz parameters (A, B, C, and M) were subsequently used to calculate the growth kinetics, including the exponential growth rate, EGR (logCFU/g/h)=BC/e; the lag-phase duration, LPD (h)=M−1/B; the generation time, GT (h)=log(2)e/BC; and the maximum population density, MPD (logCFU/g)=A+C.
Statistical analysisAn analysis of variance (ANOVA) was performed to compare the growth parameters of E. coli O157:H7 obtained from the growth curves. The ANOVA and least significance difference (LSD) between the means were calculated at 95% confidence level using the SYSTAT software. The means that differed from the calculated LSD were considered statistically significant (α=0.05).
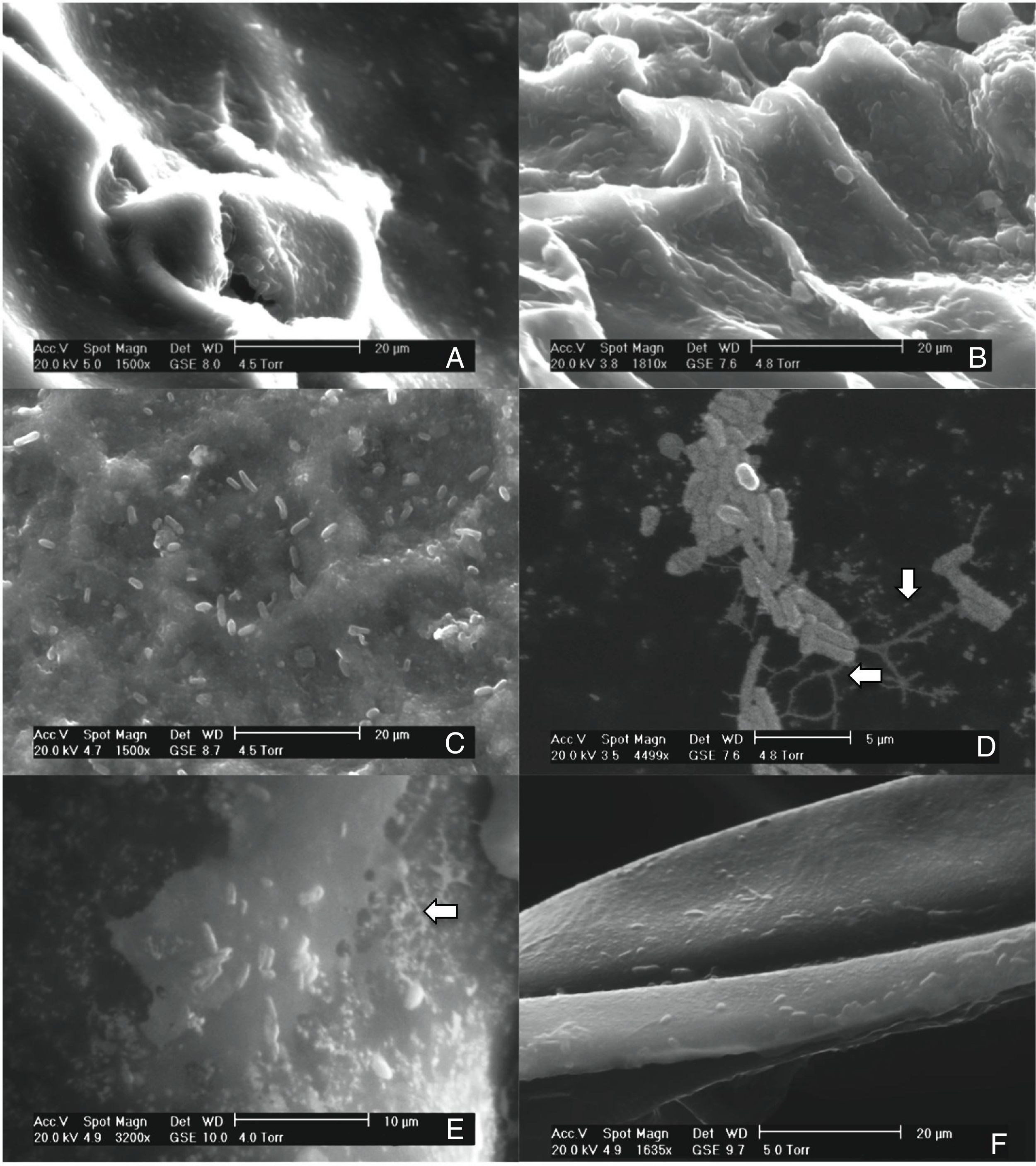
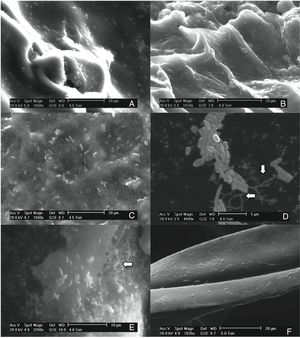
Results and discussionThis study investigated the quantification of the growth of E. coli O157:H7 on vegetables and fruits. Although the major disease outbreaks attributed to this pathogen have been associated with leafy vegetables, the results of this study showed that E. coli O157:H7 is also capable of adhering to vegetables with different physicochemical characteristics (Fig. 1). The methodology used in this study for the adhesion of E. coli O157:H7 was validated through internal controls. The bacterial cell that adhered to the surface of the samples formed a matrix with the epidermal layer (biofilm), which provided them adequate support to counter the effects of washing. E. coli O157:H7 strain possesses several fimbrial and nonfimbrial adhesins which usually do not play any role in its pathogenesis, but are involved with its adhesion to surfaces.31 For example, curli-expressing thin-aggregative fimbriae are responsible for binding to eukaryotic extracellular matrix proteins and promoting the formation of E. coli O157:H7 biofilms on inert surfaces.32,33 According to Torres et al.,31 the formation of biofilms increases the probability of survival of E. coli O157:H7, and hence, facilitates its protection against adverse environmental conditions. The samples from the vegetables and fruits that were not subjected to the final wash prior to plating (internal control) recorded higher bacterial concentration than those that were washed, suggesting the presence of both adhered and free microorganism on the surface of the samples. The SEM images of the samples obtained after the final washing clearly showed a layer of capsule bound to the cell wall of the samples that could possibly be associated with the exopolysaccharides secreted by the microorganism to form the matrix structure (Fig. 1D and E).
Scanning electron micrographs demonstrating the adhesion of Escherichia coli O157:H7 on the epicarp of vegetables and fruits, (A) beet, Beta vulgaris subsp. vulgaris (1550×); (B) habanero Chili, Capsicum chinense L., (1810×); (C) tomato, Lycopersicon esculentum (1500×); (D) radish, Raphanus sativus (4499×); (E) cucumber, Cucumis sativus (3200×); (F) onion, Allium cepa L. (1635×).
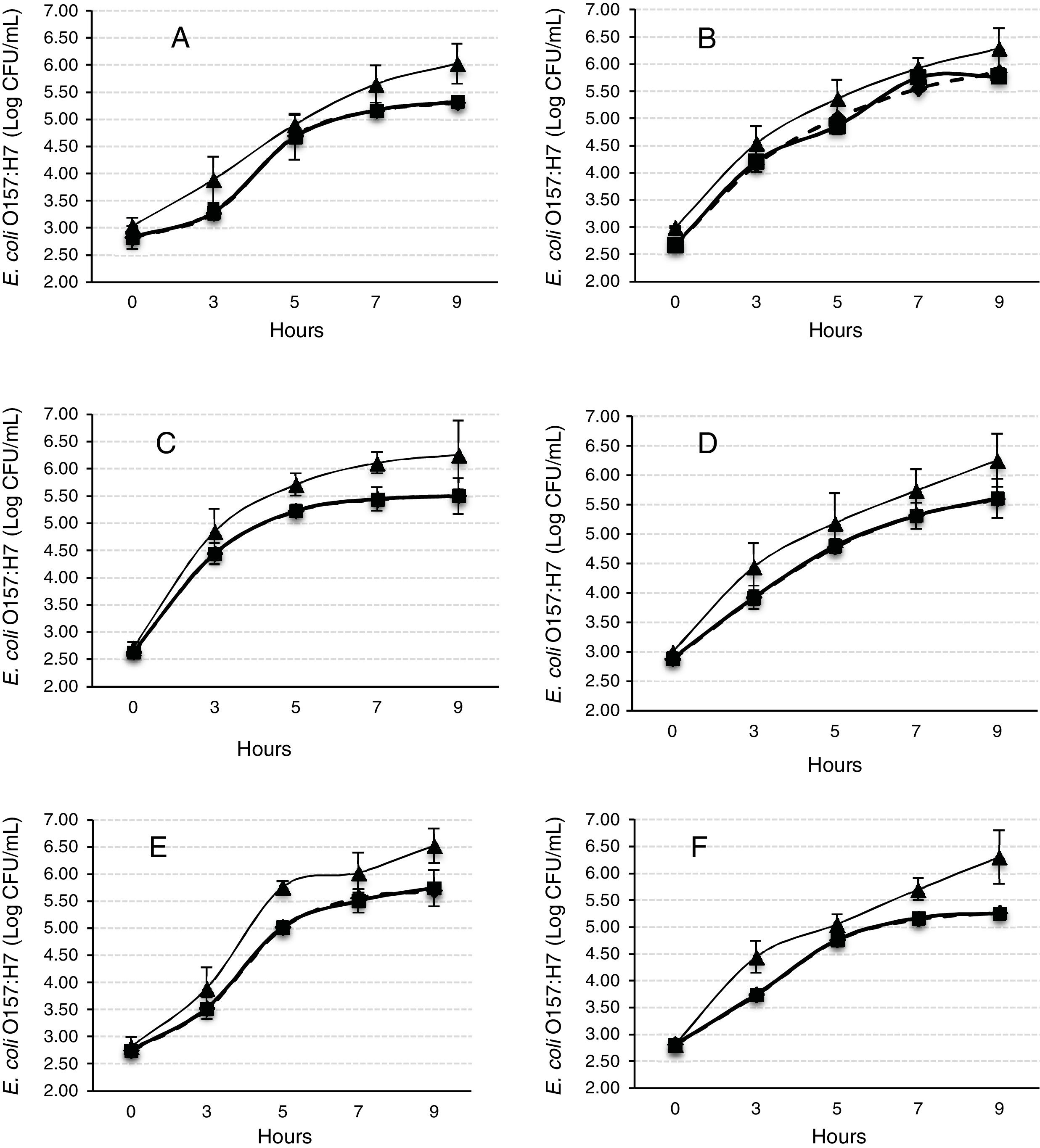
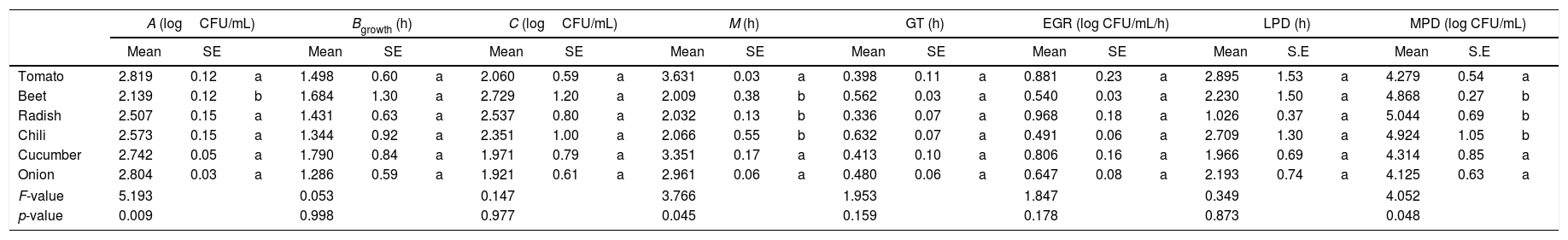
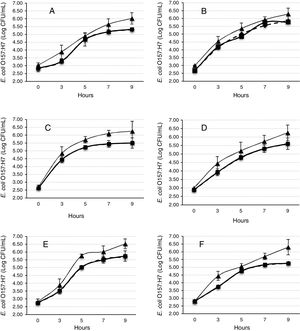
The Gompertz function fits well for the observed data with the sum of squares being less than 1 in all the cases (Fig. 2). Despite of the different intrinsic factors of the vegetables and fruits used in this study that could influence the growth of human pathogenic microorganisms, no significant difference was found in the lag-phase duration (LPD=2.17±1.03h; mean±S.E.), generation time (GT=0.47±0.07h), or exponential growth rate (EGR=0.72±0.12logCFUmL/h) of the E. coli O157:H7 adhered to the samples (Table 1). Additional information on the proximate composition of the vegetables and fruits under investigation are shown in Table 2. These values are comparable to those reported by Kim et al.,34 for perilla leaves inoculated with E. coli O157:H7 at 36°C (LPD=2.40±0.32h; EGR=0.55±0.11CFU/mL/h). The growth of the E. coli O157:H7 strain on the epicarp of the vegetables and fruits was slower than that observed under controlled laboratory conditions. The growth model of Buchanan and Klawitter,35 which considered the same temperature and pH conditions employed in this study (35±1°C; pH 6.0) displayed faster kinetic values than those observed in this study (LPD=1.54h; GT=0.33h; and EGR=0.92logCFU/mL/h). This could be an outcome of the variation in the interactions that occur between the substrate and pathogen which might not necessarily occur in a growth medium. Similarly, the duration of the lag phase is dependent on a wide variety of factors, including the initial concentration of the inoculum, the time required to recover from the physical damage or shock due to the transfer, and the time required for the synthesis of essential coenzymes or division factors and other enzymes involved in the metabolism of the substrates present in the medium.36 In general when pathogens colonize plant surfaces, they are required to evade various defense mechanisms of the host plants. Plants possess a variety of complex immune responses, which enable them to detect and initiate counter measures to control undesired proliferation of foreign substances.37 In addition, ethylene, a plant hormone, plays a vital role in the defense against plant pathogens.38 Many microorganisms considerably increase the expression of ethylene; however the transcription level of ethylene induced by E. coli O157 were ten-fold lower, suggesting that either the plants do not sense the presence of the microorganism or that the microorganism evade the defense mechanisms of the plants consequently inhibiting the activation of ethylene.39
Mathematical modeling of the growth of E. coli O157:H7 adhered to the epicarp of vegetables and fruits. The Gompertz (A, B, C, M) and derived parameters (GT, generation time; EGR, exponential growth rate; LPD, lag-phase duration; MPD, maximum population density) are shown. Data are presented as mean±standard error (n=3). The letters between the columns indicate significant differences (p<0.05).
| A (logCFU/mL) | Bgrowth (h) | C (logCFU/mL) | M (h) | GT (h) | EGR (log CFU/mL/h) | LPD (h) | MPD (log CFU/mL) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | Mean | SE | Mean | SE | Mean | SE | Mean | S.E | Mean | S.E | |||||||||
| Tomato | 2.819 | 0.12 | a | 1.498 | 0.60 | a | 2.060 | 0.59 | a | 3.631 | 0.03 | a | 0.398 | 0.11 | a | 0.881 | 0.23 | a | 2.895 | 1.53 | a | 4.279 | 0.54 | a |
| Beet | 2.139 | 0.12 | b | 1.684 | 1.30 | a | 2.729 | 1.20 | a | 2.009 | 0.38 | b | 0.562 | 0.03 | a | 0.540 | 0.03 | a | 2.230 | 1.50 | a | 4.868 | 0.27 | b |
| Radish | 2.507 | 0.15 | a | 1.431 | 0.63 | a | 2.537 | 0.80 | a | 2.032 | 0.13 | b | 0.336 | 0.07 | a | 0.968 | 0.18 | a | 1.026 | 0.37 | a | 5.044 | 0.69 | b |
| Chili | 2.573 | 0.15 | a | 1.344 | 0.92 | a | 2.351 | 1.00 | a | 2.066 | 0.55 | b | 0.632 | 0.07 | a | 0.491 | 0.06 | a | 2.709 | 1.30 | a | 4.924 | 1.05 | b |
| Cucumber | 2.742 | 0.05 | a | 1.790 | 0.84 | a | 1.971 | 0.79 | a | 3.351 | 0.17 | a | 0.413 | 0.10 | a | 0.806 | 0.16 | a | 1.966 | 0.69 | a | 4.314 | 0.85 | a |
| Onion | 2.804 | 0.03 | a | 1.286 | 0.59 | a | 1.921 | 0.61 | a | 2.961 | 0.06 | a | 0.480 | 0.06 | a | 0.647 | 0.08 | a | 2.193 | 0.74 | a | 4.125 | 0.63 | a |
| F-value | 5.193 | 0.053 | 0.147 | 3.766 | 1.953 | 1.847 | 0.349 | 4.052 | ||||||||||||||||
| p-value | 0.009 | 0.998 | 0.977 | 0.045 | 0.159 | 0.178 | 0.873 | 0.048 | ||||||||||||||||
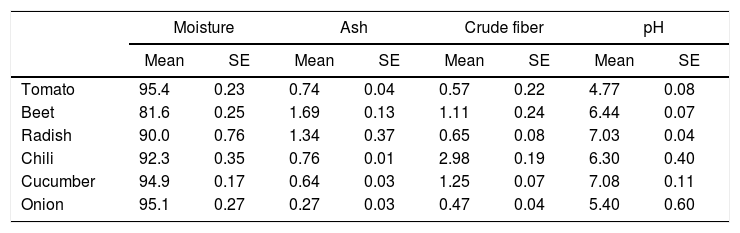
Epicarp proximal composition (%) of vegetables and fruits (n=5).
| Moisture | Ash | Crude fiber | pH | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | Mean | SE | |
| Tomato | 95.4 | 0.23 | 0.74 | 0.04 | 0.57 | 0.22 | 4.77 | 0.08 |
| Beet | 81.6 | 0.25 | 1.69 | 0.13 | 1.11 | 0.24 | 6.44 | 0.07 |
| Radish | 90.0 | 0.76 | 1.34 | 0.37 | 0.65 | 0.08 | 7.03 | 0.04 |
| Chili | 92.3 | 0.35 | 0.76 | 0.01 | 2.98 | 0.19 | 6.30 | 0.40 |
| Cucumber | 94.9 | 0.17 | 0.64 | 0.03 | 1.25 | 0.07 | 7.08 | 0.11 |
| Onion | 95.1 | 0.27 | 0.27 | 0.03 | 0.47 | 0.04 | 5.40 | 0.60 |
E. coli O157:H7 growth was lower in onion than other samples suggesting an interaction that is not completely favorable between the microorganism and the epicarp of the vegetable. In the case of tomato and cucumber, the time for the microorganism to reach the maximum growth rate (M) was significantly longer than that observed for other samples; the maximum population densities (MPD) for these vegetables were also the lowest (Table 1). The variation in the size of the bacterial populations among the different species has been attributed to a range of plant characteristics, including the leaf water content, pH, and the presence of bacterial inhibitory compounds. In addition, it occurs only when the water pressure on the surface of the product overcomes the internal gas pressure, and also it depends on the hydrophobic nature of the surface.40–42E. coli grows in a broad pH range of 4.4–10.0 with an optimum pH of 6–7.43 Specifically, E. coli O157:H7 can tolerate harsh acidic conditions, and few other strains have been reported to be able to survive even at pH 2.5–3.0 for over 4h.44 Arnold and Kaspar,45 found that E. coli O157:H7 becomes more tolerant to acid when it is in the stationary growth phase or is starved during the log-phase of growth. The systemic pH of the vegetables and fruits used in this study is in a suitable range suitable for the growth of the pathogenic microorganism (Table 2), tomato and onion possess the lowest pH (4.77 and 5.40, respectively). This suggests that the pH in tomatoes might have influenced the growth rate of E. coli O157:H7 during the log-phase, as the pathogen took longer to reach the maximum growth rate (M) in tomatoes (Fig. 2, Table 1). This finding is in agreement with a study by Beuchat46 with ripe tomatoes, which reported that the pH range of 3.9–4.4 present in the ripe tomatoes prevents or retards bacterial growth. Del Rosario and Beuchat47 also suggested that vegetables and fruit juices with a pH value less than 4.0 are, in general, not good substrates to support the growth of E. coli O157:H7.
Other researchers have shown that MPD decreases significantly when the concentration of preservatives increases or when natural microbial compounds are present.48 As in the case of cucumber the volatile oils and the methanolic and dichloromethane extracted from the peel of C. sativus exhibited antimicrobial activity.49 Cho et al.,50 reported that the high content of the volatiles such as 2,6-nonadienal and 2-nonenal show strong activity against E. coli O157:H7. In contrast to this, there are no reports on the presence of antimicrobial compounds against human pathogens in the skin of tomato and onion. Although not specifically against E. coli O157:H7, but still some studies have reported antimicrobial activity of onion aqueous extracts against gram-negative microorganisms.51 However, Rounds et al.,52 showed that powdered onion induced the growth of E. coli by 1.2logCFU/g, indicating that onion, like other vegetables, provides nutrients that enhance the growth of bacteria. Nevertheless, there is no conclusive explanation on the delay of the growth rate of E. coli O157:H7 on those vegetables.
In conclusion, the predictive models presented in this study offer a good comparative account of the growth of E. coli O157:H7 on samples with different intrinsic factors demonstrating low selectivity of the pathogen. The data suggest that selectivity of the bacterium, E. coli O157:H7 strain SS11 slightly depends on the intrinsic factors of the vegetables and fruits under study, with the clear exception of tomato, cucumber and onion, wherein the delay in the growth of the pathogen was significant. However, it is important to note that the growth models constructed under controlled environmental conditions offer limited insights and may vary widely from the observed profile of vegetables and fruits in the field, since this study was designed to inoculate the samples with E. coli O157:H7 without the presence of competitive microflora on the surface. A lower concentration of the microorganism could also be expected in the field than that was employed in the study in the laboratory. The results reported by Brackett,52 suggested that reducing the native microbial populations by washing and sanitizing or by controlled atmosphere storage, enables proliferation of human pathogens on the surface of vegetables. The reduction in the surface populations reduces competition for space and nutrients thereby providing growth potential for pathogenic contaminants. In theory, this scenario can result in an unspoiled product that is unsafe for consumption. Future research in this field should aim to verify the potential for adhesion and internalization of E. coli O157:H7 in the tissues of vegetables under realistic field conditions.
Conflicts of interestThe authors declare no conflicts of interest.
The authors are grateful to Ana Ruth Christopher Ramos from CINVESTAV-Mérida (México) for technical assistance with the scanning electron microscopy. We also thank Daniela A. Acevedo Ojeda from UMM for assistance with PCR analysis. This project was supported by FOMIX-CONACYT – Gobierno del Estado de Yucatán (México), grant no. 108103.