Gastroenteritis is one of the most common diseases during childhood, with norovirus (NoV) and sapovirus (SaV) being two of its main causes. This study reports for the first time the incidence of these viruses in hospitalized children with and without gastroenteritis in São Luís, Maranhão. A total of 136 fecal samples were tested by enzyme immunoassays (EIA) for the detection of NoV and by reverse transcription-polymerase chain reaction (RT-PCR) for detection of both NoV and SaV. Positive samples for both agents were subjected to sequencing. The overall frequency of NoV as detected by EIA and RT-PCR was 17.6% (24/136) and 32.6% (15/46), respectively in diarrheic patients and 10.0% (9/90) in non-diarrheic patients (p<0.01). Of the diarrheic patients, 17% had fever, vomiting and anorexia, and 13% developed fever, vomiting and abdominal pain. Of the 24 NoV-positive samples, 50% (12/24) were sequenced and classified as genotypes GII.3 (n=1), GII.4 (6), GII.5 (1), GII.7 (2), GII.12 (1) and GII.16 (1). SaV frequency was 9.8% (11/112), with 22.6% (7/31) in diarrheic patients and 4.9% (4/81) in nondiarrheic (p=0.04) ones. In diarrheic cases, 27.3% had fever, vomiting and anorexia, whereas 18.2% had fever, anorexia and abdominal pain. One SaV-positive sample was sequenced and classified as GII.1. These results show a high genetic diversity of NoV and higher prevalence of NoV compared to SaV. Our data highlight the importance of NoV and SaV as enteropathogens in São Luís, Maranhão.
Acute gastroenteritis (AG) is considered to be the second leading cause of death due to infection1 mainly in children under five years old, leading to nearly 760,000 deaths each year, with 75% of episodes caused by viruses.2–4
Among the main causes of viral gastroenteritis, two members of the Caliciviridae family are epidemiologically relevant: norovirus (NoV) and sapovirus (SaV). The relevance of NoV as a major cause of AG outbreaks has already been established,5 as well as its role in episodes of diarrhea that require hospitalization or visits to outpatient clinics.6 SaV are predominantly related to sporadic episodes and outbreaks of diarrhea in indoors environments, such as daycare centers, and are also associated with AG at different ages.7
NoV and SaV are non-enveloped viruses with diameters of 27–40nm, and both are composed of a single-stranded RNA genome. The NoV is classified into six genogroups, with several genotypes in the GI, GII and GIV genogroups known to cause AG in humans.8,9 GII.4 is currently regarded as the most frequent genotype, being related to approximately 70% of outbreaks, and the global epidemics that have occurred since 1995 have been caused by seven GII.4 variants.9–11 The SaV genus is divided into five genogroups, four of which can infect humans (GI-II, GIV and GV), with GI described as the most common.8,12 The transmission of both viruses occurs primarily by the fecal-oral route through direct interpersonal contact, ingestion of contaminated food and water and possibly by aerosolized particles from vomiting.13 It is believed that for NoV, fresh foods such as fruits are those most likely to be contaminated, and this probably occurs during irrigation.6
Cases of asymptomatic infection vary from 12% to 30%, which may represent a transmission facilitator.14,15 The period of incubation of these pathogens ranges from 24 to 48h, with symptoms persisting from 12 to 72h. The clinical manifestations vary from severe to moderate, mainly in NoV cases, with diarrhea, vomiting, nausea, and fever that sometimes lead to dehydration, particularly in children and the elderly, which may lead to fatal outcomes.16
Research conducted in hospitals, clinics and communities in several locations in Brazil showed NoV occurrence ranging from 8.6% to 39.7%.15,17–20 However, studies involving these viruses are scarce in the north and northeast regions.
In this study, we sought to determine the prevalence of and partially characterize NoV and SaV in hospitalized children with or without diarrhea in the city of São Luís, Maranhão, Brazil, during the period from 1997 to 1999.
Materials and methodsLocation of study and patientsA total of 250 stool samples were obtained from children less than three years old who were hospitalized with diarrhea or other symptoms at the Materno Infantil Unit at the Universitary Hospital (HUMI) of the Federal University of Maranhão (UFMA) from June 1997 to June 1999. Previously, these samples were tested for rotavirus (RV)21 and astrovirus (AstV),22 and only the samples with negative results for both viruses were used in the present study. All samples were stored at −20°C, and 136 specimens (46 diarrheic and 90 without diarrhea) were selected for this research. This study was approved by the Evandro Chagas Institute Ethical Committee under the number 284.852.
Fecal suspensions and enzyme immunoassay (EIA)Fecal suspensions were prepared at a dilution of 10% (weight/volume) and were tested for the presence of NoV using a commercial enzyme immunoassay (EIA) (3rd generation RIDASCREEN® Norovirus EIA, R-Biopharm, Darmstadt, Germany) according to the manufacturer's recommendations.
Viral RNA extraction and reverse transcription (RT)A volume of 300μl of fecal suspension was used for nucleic acid extraction by the silica method23 with a final volume of 45μl that was stored at −20°C until use. A volume of 6μl of RNA was used to obtain the complementary DNA (cDNA) by reverse transcription (RT) using a random primer.
Molecular detectionThe specimens were screened by polymerase chain reaction (PCR) with the primers Mon431/Mon433 and Mon432/Mon434, which are specific for the NoV RNA polymerase sequences of genogroups I and II, respectively.24 Samples that tested negative with this pool of primers were also tested with primers P289/P290 for the detection of SaV.25
All NoV-positive samples were subjected to two other PCR analyses: the first one amplified part of the viral capsid region of NoV (Cap A, B2, B1 and Cap C, D3, D1)26 and the other amplified the B (polymerase) and C (capsid) junction regions (primers G2SKR and Mon/432).27
Purification and sequencing of RT-PCR calicivirus ampliconsThe samples subjected to nucleotide sequencing were purified with the QIAquick® PCR Purification (Qiagen) and QIAquick® Gel Extraction (Qiagen) commercial kits according to the manufacturer's instructions and sequenced using the Big Dye Terminator kit (v.3.1) (Applied Biosystems) in an automatic sequencer. The analysis and editing of the chromatograms were performed by the BioEdit Sequence Alignment Editor (v. 7.0) program and compared to other sequences stored in GenBank with the BLAST program. Phylogenetic grouping was constructed using maximum likelihood analysis. The evolutionary model selected for the polymerase tree was TPM3+G4 and for the capsid tree was the TIM2e+G4. For both trees, testing was performed with 1000 bootstrap replicates, and the cut-off value was 70%.
Statistical analysisStatistical analysis was performed using BioEstat 5.0 software.28 The odds ratio test was used to correlate infection with age ranges. The chi-square test and Fisher's exact test were used to determine whether the results for the two groups (NoV and SaV) were statistically significant. Simple regression analysis was used to relate each symptom to NoV and SaV infections.
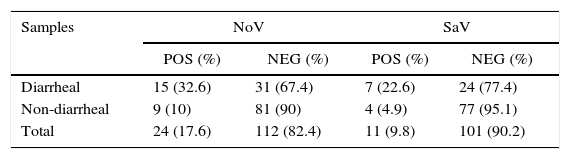
ResultsThe NoV frequency was 17.6% (24/136), with 75% (18/24) detected only by PCR and 25% (6/24) detected by both PCR and EIE. For SaV, the frequency was 9.8% (11/112). Of the two groups studied (i.e., with and without diarrhea), the first displayed a higher incidence of infection for both viruses: NoV (32.6% – p<0.012) and SaV (22.6% – p<0.04) (Table 1).
Detection of norovirus and sapovirus in fecal samples collected from hospitalized children with and without diarrhea from São Luís, Maranhão from June 1997 to June 1999.
| Samples | NoV | SaV | ||
|---|---|---|---|---|
| POS (%) | NEG (%) | POS (%) | NEG (%) | |
| Diarrheal | 15 (32.6) | 31 (67.4) | 7 (22.6) | 24 (77.4) |
| Non-diarrheal | 9 (10) | 81 (90) | 4 (4.9) | 77 (95.1) |
| Total | 24 (17.6) | 112 (82.4) | 11 (9.8) | 101 (90.2) |
Of the 24 NoV-positive samples, 50% (12/24) were used to perform nucleotide sequencing, with 66.7% (8/12) from diarrheic patients and 33.3% (4/12) from the non-diarrheic group.
In the diarrheic group, three samples were characterized as genotype GII.4 and five as genotypes GII.3, GII.5, GII.7, GII.12 and GII.16 (one each). In the non-diarrheic group, two samples were characterized as GII.4 and two as GII.7.
Of the 12 samples characterized, four were sequenced with primers targeting two regions: region B (RdRp) and region D (viral capsid), with the genotypes confirmed in both regions as GII.3 and GII.16 and two as GII.4. Two samples were sequenced using the junction region and were classified as genotypes GII.4 and GII.5. Five other samples were characterized by the RdRp region B, with three classified as GII.P4 and two as GII.P7. One sample was characterized as GII.12 by region D of the viral capsid (Table 2 and Figs. 1 and 2).
Distribution of norovirus genotypes detected in fecal samples collected from hospitalized children with and without diarrhea in São Luís, Maranhão from June 1997 to June 1999.
| Samples | Nucleotide sequencing | Total | |||
|---|---|---|---|---|---|
| Only region B – RdRp | Only region D – viral capsid | Pos both regions | Junction region | ||
| Diarrheal | GII.P4 (2) GII.P7 | GII.12 | GII.3 GII.16 GII.4 | GII.5 | 8 |
| Non-diarrheal | GII.P4 GII.P7 | GII.4 | GII.4 | 4 | |
| Total | 5 | 1 | 4 | 2 | 12 |
Phylogenetic tree based on the polymerase sequence of norovirus. The study samples are marked in bold, and the tree was constructed using maximum likelihood analysis. The evolutionary model selected for the polymerase tree was TPM3+G4. The test was performed with 1000 bootstrap replicates, and the cut-off value was 70%.
Phylogenetic tree based on the capsid region of norovirus. The study samples are marked in bold, and the tree was constructed using maximum likelihood analysis. The evolutionary model selected for the polymerase tree was TIM2e+G4, and the test was performed with 1000 bootstrap replicates, and the cut-off value was 70%.
No positive samples were characterized as NoV genogroup I. One diarrheic sample was sequenced for SaV and classified as GII.1.
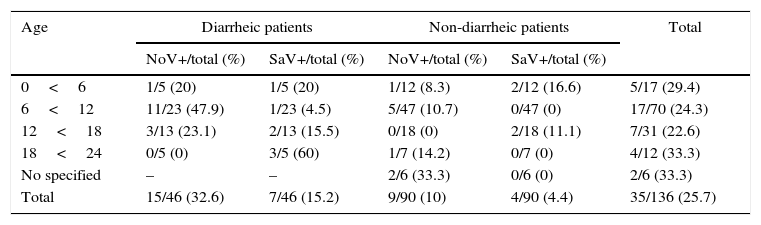
The highest NoV frequency according to age groups was found between 6 and 12 months of age (OD=1.7%, p<0.2) and for SaV frequency, between 18 months to two years of age (Table 3).
Frequency by age of positive cases for norovirus and sapovirus in hospitalized children with and without acute gastroenteritis, June 1997 to June 1999 in São Luís, Maranhão.
| Age | Diarrheic patients | Non-diarrheic patients | Total | ||
|---|---|---|---|---|---|
| NoV+/total (%) | SaV+/total (%) | NoV+/total (%) | SaV+/total (%) | ||
| 0<6 | 1/5 (20) | 1/5 (20) | 1/12 (8.3) | 2/12 (16.6) | 5/17 (29.4) |
| 6<12 | 11/23 (47.9) | 1/23 (4.5) | 5/47 (10.7) | 0/47 (0) | 17/70 (24.3) |
| 12<18 | 3/13 (23.1) | 2/13 (15.5) | 0/18 (0) | 2/18 (11.1) | 7/31 (22.6) |
| 18<24 | 0/5 (0) | 3/5 (60) | 1/7 (14.2) | 0/7 (0) | 4/12 (33.3) |
| No specified | – | – | 2/6 (33.3) | 0/6 (0) | 2/6 (33.3) |
| Total | 15/46 (32.6) | 7/46 (15.2) | 9/90 (10) | 4/90 (4.4) | 35/136 (25.7) |
In addition to diarrhea, the other four clinical symptoms (fever, vomiting, anorexia and abdominal pain) in the symptomatic patients were analyzed. Fever was the only symptom that showed a statistically significant association (p<0.04). Of the positive cases for NoV, 28.6% had fever, vomiting and anorexia, and 21.6% had fever, vomiting and abdominal pain. Of the cases positive for SaV, 42.8% were associated with fever, vomiting and anorexia, and 28.6% with fever, anorexia and abdominal pain. The analysis of the symptoms of NoV- and SaV-positive cases is shown in Table 4.
Analysis of the clinical symptoms presented by children hospitalized with acute gastroenteritis positive for norovirus and sapovirus in São Luís, Maranhão, from June 1997 to June 1999.
| Symptoms | Positive cases | |
|---|---|---|
| NoV (%) | SaV (%) | |
| Vomit | 1 (7.1) | – |
| Vomit/anorexia | 1 (7.1) | – |
| Vomit/anorexia/abdominal pain | 1 (7.1) | – |
| Vomit/fever/anorexia | 4 (28.6) | 3 (42.8) |
| Vomit/fever/abdominal pain | 3 (21.6) | – |
| Vomit/fever/anorexia/abdominal pain | 2 (14.2) | 1 (14.3) |
| Fever/anorexia | 1 (7.1) | 1 (14.3) |
| Fever/anorexia/abdominal pain | 1 (7.1) | 2 (28.6) |
| Total | 14 | 7 |
In the non-diarrheic cases, nine samples were positive for NoV and four to SaV.
Currently, studies related to the circulation of these pathogens in northeastern Brazil are scarce, and this is the first study to investigate the prevalence of these viruses in São Luís, state of Maranhão.
In the present study, the prevalence of NoV infection (17.6%) in cases of hospitalized children with and without gastrointestinal infection was higher than that detected in children less than five years old in the state of Acre (north) (12.7%),29 similar to the results found in an African descendant community in Pará (north) (19.7%)30 and lower than that detected in Niterói (southeast) (30.3%),15 where most infections by NoV were recorded in children less than two years of age. For SaV, the incidence was higher (11%) than that detected in the United States (5%)31 and in Italy (3%).32
Of the total samples analyzed, the highest prevalence was observed among diarrheic patients, which was significant for both NoV (32.6% – p<0.012) and SaV (22.6% – p<0.04). These findings corroborate the results obtained for NoV in a study conducted in São Paulo (southeast) (36.2%)17 and also in Belém (north) (35.4%),33 both reporting increased frequency of this pathogen in symptomatic patients. With regard to SaV, the prevalence detected in the present study for symptomatic patients was less than that observed in India (39%).34 Detection of asymptomatic cases emphasizes that even without clinical signs, viral dispersion may be taking place, which is another source of transmission of this pathogen.
In the present study, NoV was determined by three different sets of primers, one specific for the RNA polymerase (region B, ORF1), another specific for the capsid (region D, ORF2) and the third specific for the junction region (region B, ORF1 and region C, ORF2). Nucleotide sequences allowed classification of the six samples as genotype GII.4. According to the literature, currently, strains belonging to this genogroup are showing higher frequencies in cases of acute gastroenteritis around the world, accounting for up to 80% of outbreaks worldwide and leading to the emergence of new variants every two or three years.35–37
Two samples were grouped as genotype GII.12 and GII.16. GII.12 has been associated with outbreaks in the years 2009 and 2010, with 16% of gastroenteritis in the United States38 caused by NoV and was first described in Brazil in 2009, simultaneously with GII.16.20 Our study demonstrates that these two strains were already circulating in São Luís, Maranhão in the years 1997 and 1999, respectively.
We also detected the genotypes GII.3 and GII.7. GII.3 strains are usually associated with sporadic cases of infection among breastfeeding infants, especially those younger than six months old, besides being reported together with pandemic strains of GII.4 in foodborne outbreaks.39 All samples showed negative results for NoV GI. Most studies have demonstrated higher prevalence of NoV GII, which may be related to their broad genetic diversity.40
The GII.4 genotype was predominant, in accordance with other studies that reported a higher prevalence of this genotype; however, five other types were also observed during the period of 1997–1999, showing genetic diversity among strains circulating in this population. This fact could influence the future development and implementation of a possible vaccine against NoV.
One SaV sample was sequenced and classified as GII.1. This genotype was also detected in the stool sample of a child with AG in a study carried out in Salvador, Brazil, in 200241 and described in the metropolitan region of Belém, being present in ten of the fifteen (66.7%) samples characterized.42
The age group most affected by NoV was children between six months and one year of age (47.9%). This prevalence was higher than that found in Peru (9%)43 and reinforces the data found in Burkina Faso44 and in Belém,45 where 36% and 45.1%, respectively, of children at this age were infected with NoV, while much lower frequencies were found in those that were older than two years. SaV was more prevalent among children eighteen months to two years old. These results differ from those described in Japan, where SaV was associated with children 36 months of age (91%).46 However, it is important to take into account the limited number of cases analyzed in each group in the present study.
Considering diarrhea as the inclusion criteria, this study examined the prevalence of other clinical manifestations that were present in the majority of positive cases, such as vomiting, anorexia and fever. These symptoms may be related to the clinical condition caused by NoV and SaV, as demonstrated by Nordgren et al.,44 who found an association with anorexia (62%), fever (49%) and vomiting (49%) among the positive cases for NoV. In addition to that, distress was described in some positive cases.
One limitation of the present study is that we only tested samples negative for RV and HAstV, even though co-infection involving enteric viruses is relatively common in cases of acute gastroenteritis as suggested by some reports on this matter.18,47,48 In Brazil, Victoria et al.49 reported in hospitalized children from Rio de Janeiro a rate of 4% co-infection by RV and NoV.
Although this study was conducted with samples collected in 1997–1999, our data may be useful as a reference for possible future molecular epidemiological studies (including evolutionary analyses) on NoV and SaV infections in Brazil.
ConclusionThe presence of NoV and SaV in hospitalized children in São Luís, Maranhão, reinforces that this age group is susceptible to infection by these agents, in line with general reports in the literature. The higher number of positive cases in symptomatic patients was expected, and our results also emphasize the diversity of genotypes found in the samples. These results demonstrate the circulation of NoV and SaV and indicate that more studies are necessary, considering that epidemiological information about these viruses in São Luis, Maranhão is still limited.
Conflicts of interestThe authors declare no conflicts of interest.