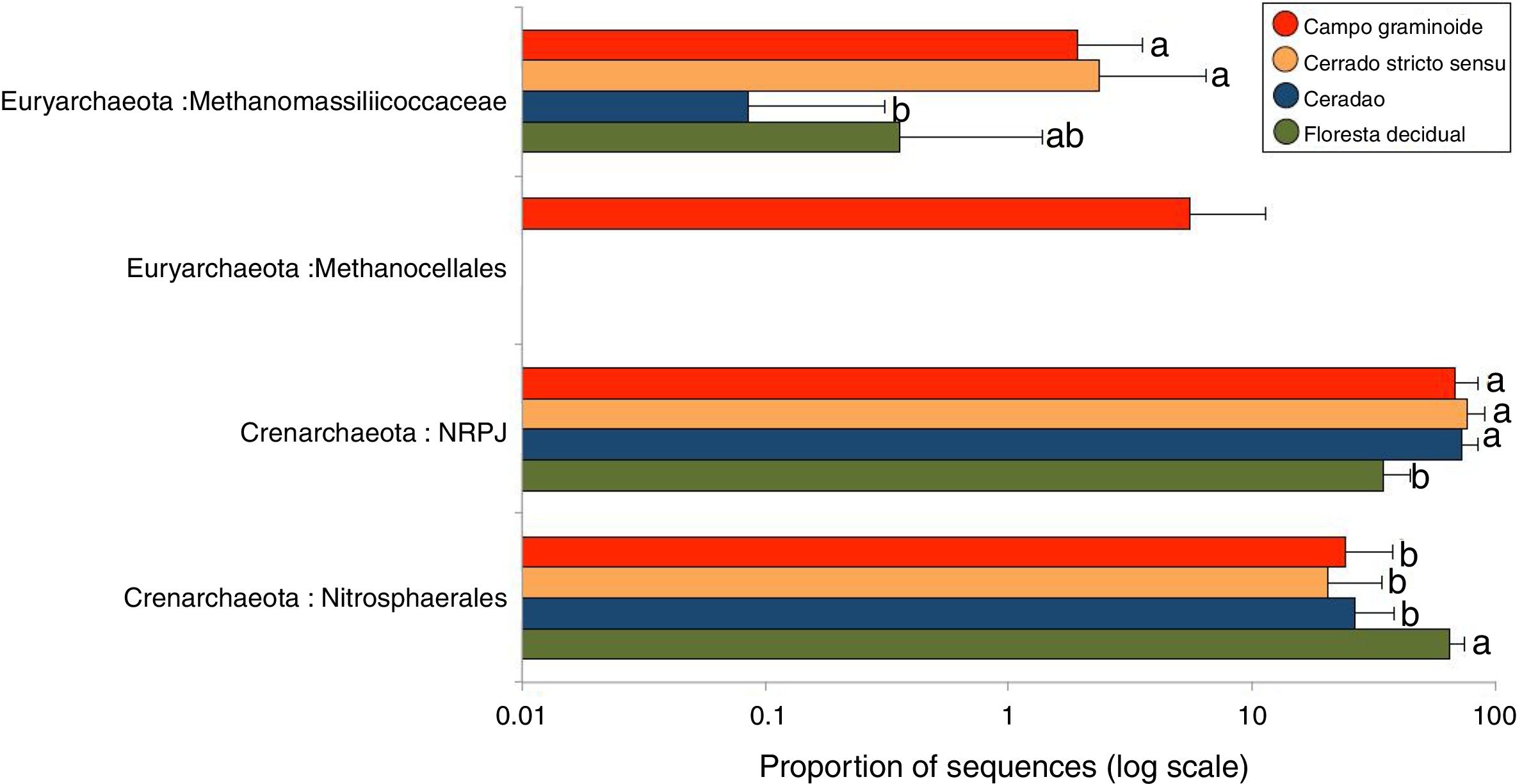
We used 16S rRNA sequencing to assess the archaeal communities across a gradient of Cerrado. The archaeal communities differed across the gradient. Crenarcheota was the most abundant phyla, with Nitrosphaerales and NRPJ as the predominant classes. Euryachaeota was also found across the Cerrado gradient, including the classes Metanocellales and Methanomassiliicoccaceae.
Soil microorganisms constitute a third of the Earth's biomass, playing important ecological and biogeochemical roles in terrestrial ecology. Among the microorganisms that inhabit soils, the domain Archaea is particularly important because it is abundant and plays important roles in C and N cycling.1 This domain constitutes one of the three major evolutionary lineages of life on Earth, and although it has long been believed that these organisms mainly inhabit extreme environments, previous studies demonstrated that they are widely distributed around the world in different types of soils.2,3
Since the recognition of the archaeal domain in the tree of life,4 studies have been focused to explore their presence and function in a wide range of environments. With the advance of DNA-based molecular tools and genomic analysis, the knowledge of this domain has evolved and the current archaeal classification is rapidly moving. Presently, 20 archaeal phyla are recognized by small subunit RNA databases; however, it has been reported that 14 phyla have no cultured representative.5 Most of culturable members of Archaea belong to the two main phyla Euryarchaeota and Crenarchaeota. Other new phyla have been proposed in the last years, for example Nanoarchaeota, based on the isolation of a hypertehermophilic symbiont Nanoarchaeum equitans6 and Thaumarchaeota.7 Most recently, two superphylum have been proposed, namely (i) TACK, which includes the phyla Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota,8 and (ii) Asgard, which consist in a sister group to TACK and are considered more closely related to the original eukaryote.9 Together with the advance of Archaea classification, their functional role in the ecosystem has also been the focus of recent studies. Members of Archaea carry out many steps in the nitrogen cycle, such as nitrate-based respiration and denitrification.10 Communities of autotrophic and heterotrophic Archaea catalyze iron and sulfur oxidation, which influence the release rate of metals and sulfur to the environment.11 Also, regarding the carbon cycle, all known methanogen organisms belong exclusively to Archaea domain and are generally found in oxygen-depleted environments.12
In Brazil, studies regarding the diversity of Archaea have been conducted for some important ecosystems, such as Amazonian and Atlantic forests,13,14 as well as in mangroves.15 In the case of the Brazilian Cerrado, previous studies investigated the diversity of archaea in soils from the Central Plateau and found that Crenarcheota was the most abundant archaeal group in the ecosystem.3,16 However, it has been hypothesized that the vegetation and soil conditions in the Cerrado from Northeastern Brazil differs from those of the Central Plateau.17 Therefore, the pattern of Archaea communities would be different because the vegetation and soil conditions drive soil microorganisms. Previous studies have shown differences between bacterial diversity in the soils obtained from Cerrado in the Northeast when compared to those from the Central Plateau.18 Therefore, we used next-generation sequencing of the 16S rRNA gene in soil samples from vegetation gradients obtained from Campo graminoide, Cerrado stricto sensu, Cerradao and Floresta decidual to understand the diversity of Archaea in the soils of Cerrado in Northeastern Brazil.
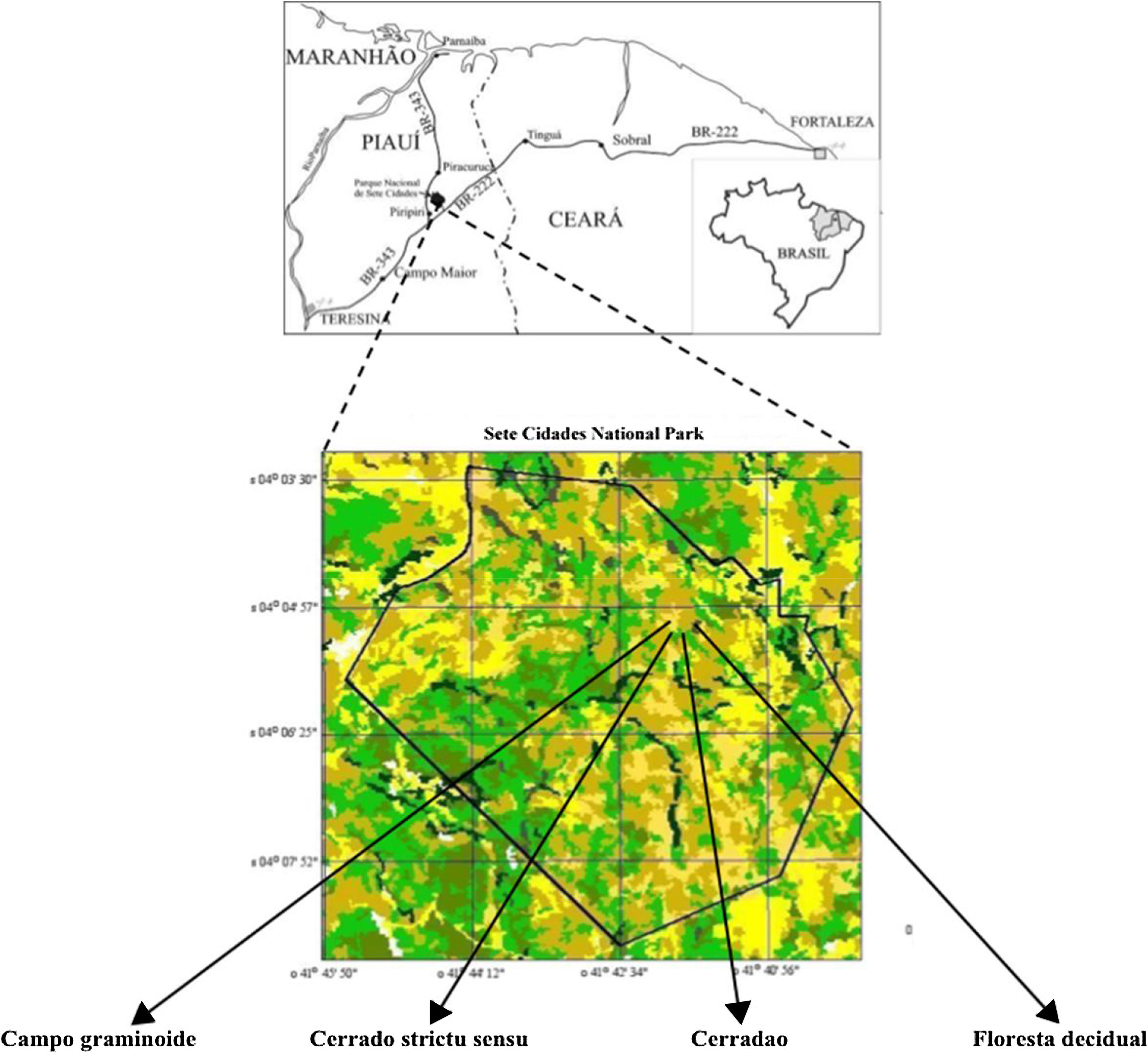
The study was conducted in a preserved Brazilian Cerrado within Sete Cidades National Park (PNSC) (04°02′-08′S and 41°40′-45′W), Northeast, Brazil. This region presents two distinct seasons (wet and dry) with an annual average temperature at 25°C and rainfall of 1558mm distributed in February through April. We evaluated four different areas (with 1000m2 each one) in a gradient of vegetation (‘Campo graminoide’, ‘Cerrado stricto sensu’, ‘Cerradao’ and ‘Floresta decidual’) (Table 1; Fig. 1).
Vegetation diversity indices in the Cerrado areas.19
| Campo graminoide | Cerrado stricto sensu | Cerradao | Floresta decidual | |
|---|---|---|---|---|
| Plant richnesse | 4.7 | 11 | 17 | 18 |
| Plant diversityf | 0.2 | 0.85 | 1.10 | 1.11 |
| Plant densityg | 4.7 | 27.1 | 35.0 | 51.8 |
| Vegetationh | a | b | c | d |
Andropogon fastigiatus; Aristida longifolia; Terminalia fagifolia; Magonia pubescens; Hymenaea courbaril; Plathymenia reticulata; Qualea grandiflora; Combretum mellifluum; Lippia origanoides; Anacardium occidentale; Simarouba versicolor; Vatairea macrocarpa.
Aspidosperma discolor; Parkia platycephala; Terminalia fagifolia; Piptadenia moniliformis; Plathymenia reticulata; Qualea parviflora; Anacardium occidentale; Copaifera coriacea; Thiloa glaucocarpa; Casearia grandiflora.
Each area was separated in three transects (replication) where soil samples were collected at top soil layer (0–20cm depth; three points per transect) in March (wet season), 2014. All soil samples were immediately stored in sealed plastic bags and transported in an icebox to the laboratory. A portion of the soil samples was stored in bags and kept at −20°C for DNA analysis and another portion was air-dried, sieved through a 2-mm screen and homogenized for chemical analyses using standard laboratory protocols.20
Soil DNA was extracted from 0.5g (total humid weight) of soil using the PowerLyzer PowerSoil DNA Isolation Kit (MoBIO Laboratories, Carlsbad, CA, USA), according to the manufacturer's instructions. The quality of the extracted DNA was checked with the Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and it was quantified by Qubit® 2.0 fluorometer using the dsDNA BR Assay kit (Invitrogen™). The integrity of the DNA was also confirmed by electrophoresis in a 0.8% agarose gel with 1× TAE buffer. The amplicon library of the archaeal 16S rRNA genes were amplified using the region-specific primers (515F/806R).21 The first step amplification comprised 25μL reaction containing the following: 14.8μL of nuclease-free water (Certified Nuclease-free, Promega, Madison, WI, USA), 2.5μL of 10× High Fidelity PCR Buffer (Invitrogen, Carlsbad, CA, USA), 1.0μL of 50mM MgSO4, 0.5μL of each primer (10μM concentration, 200pM final concentration), 1.0 unit of Platinum Taq polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), and 4.0μL of template DNA (10ng). The conditions for PCR were as follows21: 94°C for 4min to denature the DNA, with 35 cycles at 94°C for 45s, 50°C for 60s, and 72°C for 2min, with a final extension of 10min at 72°C. In the second step, a unique pair of Illumina Nextera XT indexes (Illumina, San Diego, CA) was added to both ends of the amplified products. Each 50μL reaction contained the following: 23.5μL of nuclease-free water (Certified Nuclease-free, Promega, Madison, WI, USA), 5.0μL of 10× High Fidelity PCR Buffer (Invitrogen, Carlsbad, CA, USA), 4.8μL of 25mM MgSO4, 1.5μL of dNTP (10mM each), 5.0μL of each Nextera XT index (Illumina, San Diego, CA, USA), 1.0 unit of Platinum Taq polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), and 5.0μL of each product from previous PCR. The conditions for this second round PCR were as follows: 95°C for 3min to denature the DNA, with 8 cycles at 95°C for 30s, 55°C for 30s, and 72°C for 30min, with a final extension of 5min at 72°C.
After indexing, the PCR products were cleaned up using Agencourt AMPure XP – PCR purification beads (Beckman Coulter, Brea, CA, USA), according to the manufacturer's manual, and quantified using the dsDNA BR assay Kit (Invitrogen, Carlsbad, CA, USA) on a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). Once quantified, different volumes of each library were pooled into a single tube in equimolar concentration. After quantification, the molarity of the pool was determined and diluted to 2nM, denatured, and then diluted to a final concentration of 8.0pM with a 20% PhiX (Illumina, San Diego, CA, USA) spike for loading into the Illumina MiSeq sequencing machine (Illumina, San Diego, CA, USA).
Sequence data were processed using QIIME following the UPARSE standard pipeline according to Brazilian Microbiome Project22 to produce an OTU table and a set of representative sequences. Briefly, the reads were truncated at 240bp and quality-filtered using a maximum expected error value of 0.5. Pre-filtered reads were dereplicated and singletons were removed and filtered for additional chimeras using the RDP_gold database using USEARCH 7.0. These sequences were clustered into OTUs at a 97% similarity cutoff following the UPARSE pipeline. After clustering, the sequences were aligned and taxonomically classified against the Greengenes database (version 13.8).23 The sequences were submitted to the NCBI Sequence Read Archive under the number SRP091586.
Redundancy Analysis (RDA) was used to determine the correlation between archaeal community structure and soil physicochemical properties. The OTU table was initially analyzed using Detrended Correspondence Analysis (DCA) to evaluate the gradient size of the species distribution, which indicated linearly distributed data (length of gradient <3), revealing that the best-fit model for the data was RDA. Forward selection (FS) and the Monte Carlo permutation test were applied with 1000 random permutations to verify the significance of soil chemical properties upon a microbial community. We used permutational multivariate analysis of variance (PERMANOVA)24 to test whether sample categories harbored significantly different archaeal community structure. RDA plots were generated using the software Canoco 4.5 (Biometrics, Wageningen, The Netherlands), and PERMANOVA and alpha diversity index were calculated with the software PAST.25 To determine the differences in abundance of archaeal groups among soil samples, the Statistical Analysis of Metagenomic Profiles (STAMP) software package was used.26 The OTU table was used as input, and P-values were calculated using the two-sided Welch's t-test,27 while confidence intervals were calculated using DP Welch's inverted and correction was made using Benjamini–Hochberg false discovery rate.28
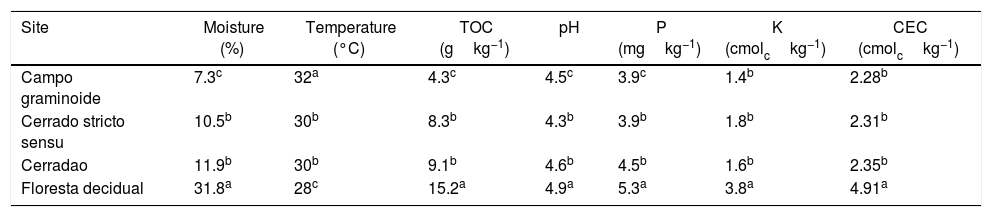
In this study, we examined the archaeal structure and composition in the soils from vegetation gradients from the Cerrado. We used 16S rRNA sequencing to assess the archaeal communities and correlated them with soil chemical parameters. Our results showed that soil parameters differed between the sampling sites. Areas under Campo graminoide, Cerrado stricto sensu and Cerradao presented the highest soil temperature and the lowest N, P, K, and CEC compared with Floresta decidual (Table 2). Soil moisture, pH, and TOC content were similar between Cerrado stricto sensu and Cerradao, and they were the lowest and highest in Campo graminoide and Floresta decidual, respectively.
Soil physicochemical properties at different sites across the gradient of Cerrado.
| Site | Moisture (%) | Temperature (°C) | TOC (gkg−1) | pH | P (mgkg−1) | K (cmolckg−1) | CEC (cmolckg−1) |
|---|---|---|---|---|---|---|---|
| Campo graminoide | 7.3c | 32a | 4.3c | 4.5c | 3.9c | 1.4b | 2.28b |
| Cerrado stricto sensu | 10.5b | 30b | 8.3b | 4.3b | 3.9b | 1.8b | 2.31b |
| Cerradao | 11.9b | 30b | 9.1b | 4.6b | 4.5b | 1.6b | 2.35b |
| Floresta decidual | 31.8a | 28c | 15.2a | 4.9a | 5.3a | 3.8a | 4.91a |
TOC, total organic C; CEC, cation exchange capacity.
Values followed by the same letter within each column are not significantly different at the 5% level, as determined by Student's t-test.
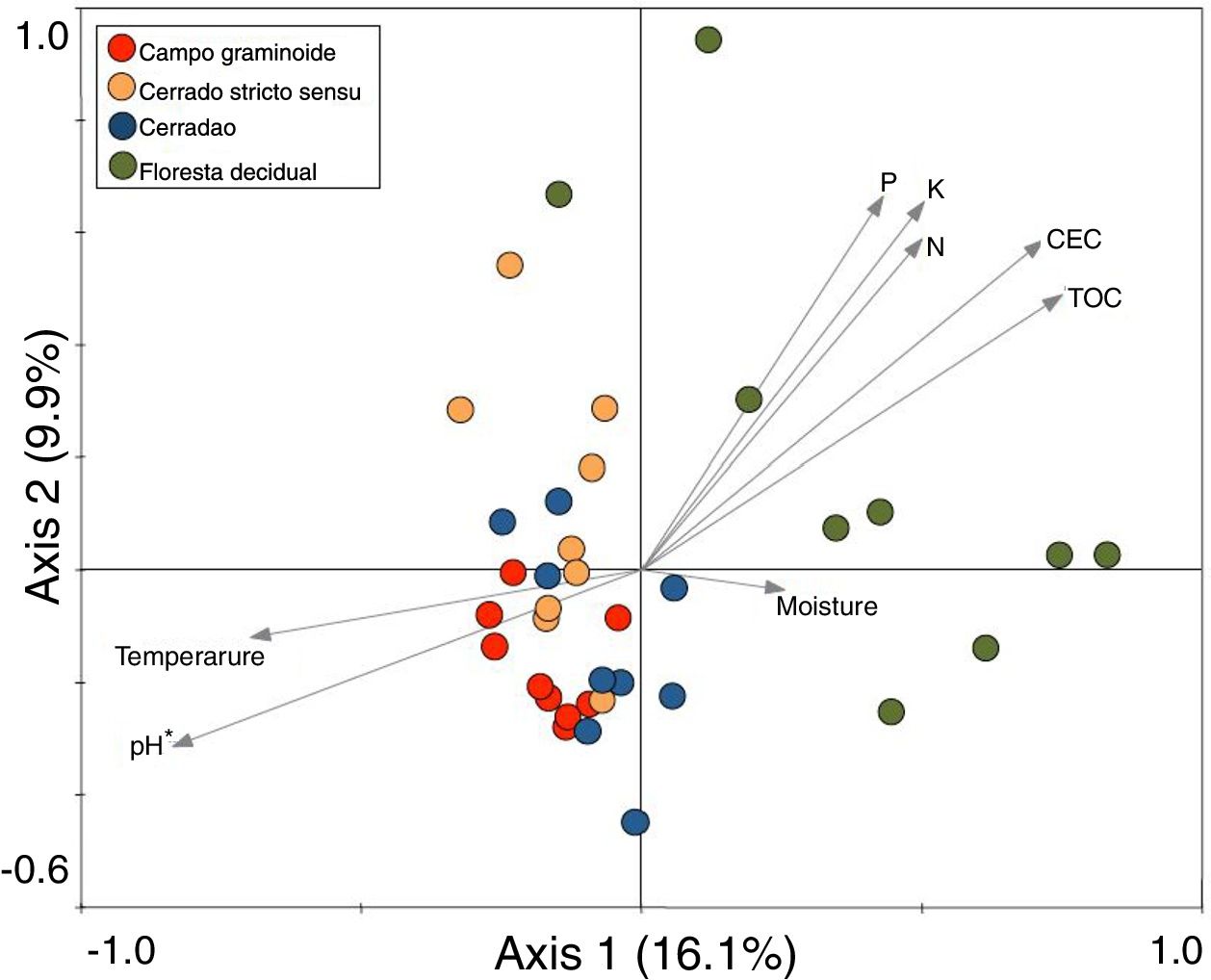
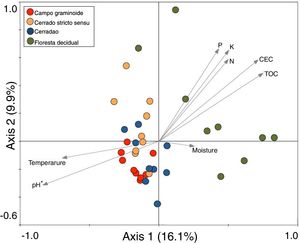
These markedly different soil conditions, mainly between Campo graminoide and Floresta decidual, exist because of the differences in plant cover and the communities, both of which can influence soil properties. For example, Campo graminoide is covered mostly by grasses, whereas Floresta decidual is covered by trees. RDA showed the differences between the archaeal communities across the gradients (Fig. 2) and indicated that the Floresta decidual was the most distinct compared to the other areas.
Redundancy analysis (RDA) of archaeal community patterns and soil characteristics from samples of Campo graminoide, Cerrado stricto sensu, Cerradao and Floresta decidual. Arrows indicate correlation between environmental parameters and archaeal profile. The significance of these correlations were evaluated via the Monte Carlo permutation test and is indicated by * (P<0.05).
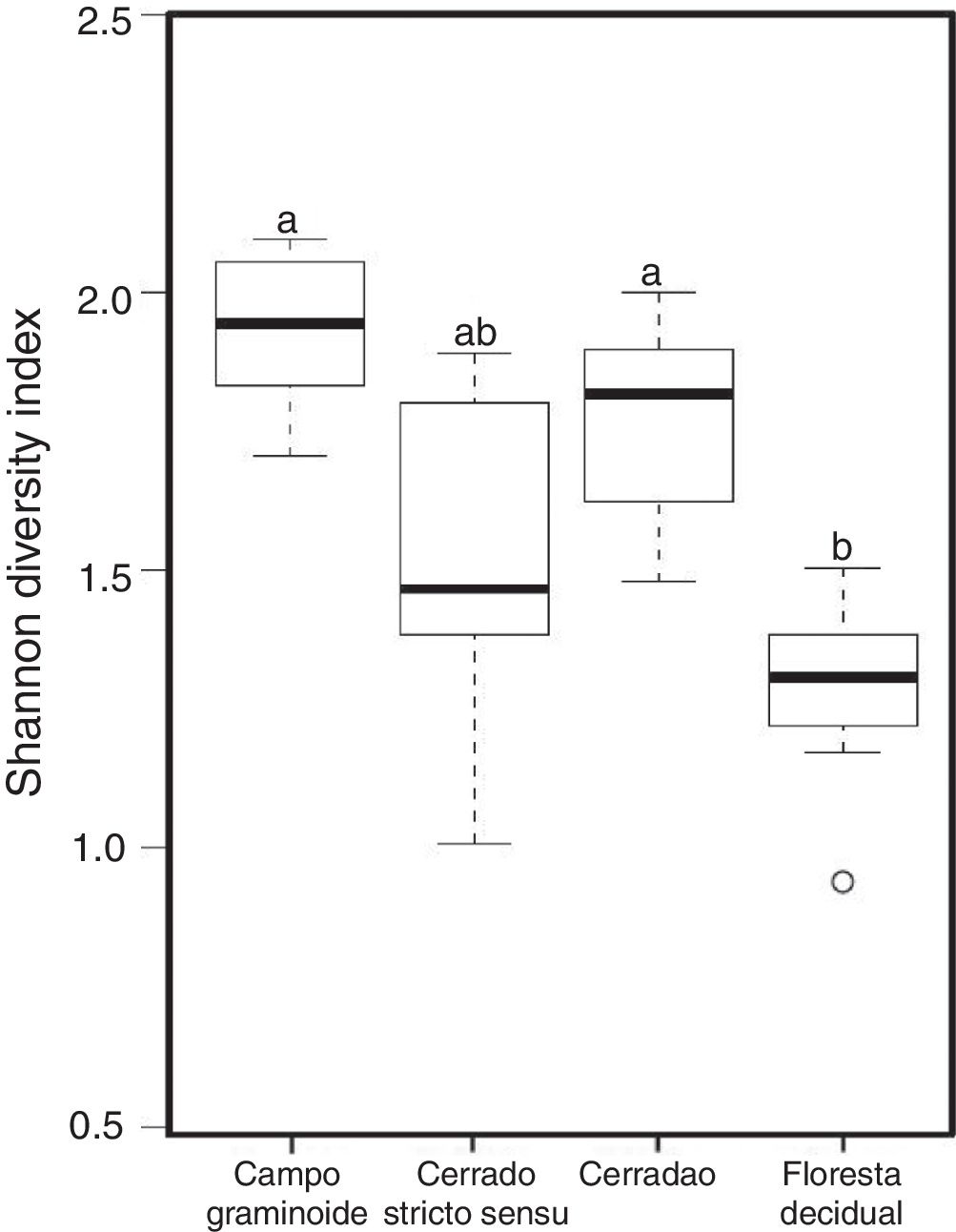
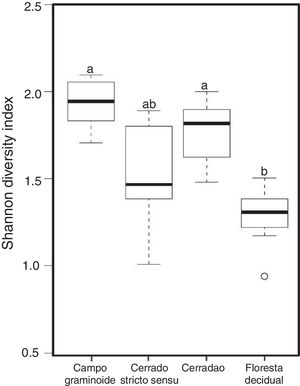
Additionally, RDA showed that the composition of archaea communities was strongly correlated with specific soil properties across the Cerrado gradients. Our results showed that the archaeal community of Floresta decidual correlated with soil moisture, TOC, CEC, P, K, and N. On the other hand, soil temperature and pH correlated with Campo graminoide, Cerrado stricto sensu and Cerradao. These results indicate that the archaea community differs across the gradients of the Cerrado and is influenced by soil conditions. Because Campo graminoide, Cerrado stricto sensu and Cerradao had similar soil conditions, their archaea communities were more similar to each other when compared to Floresta decidual. According to Catão et al.3 the soil conditions drive the differences in the diversity of Archaea in the Brazilian Cerrado. These authors compared and contrasted Cerrado denso and Mata de Galeria from the Brazilian Cerrado and found differences in the community structures of Archaea. The varied soil conditions found across the gradients of vegetation have influenced the bacterial diversity.18 Interestingly, our results highlight the soil pH as a significant driver of archaea communities in these soils. According to Gorres,29 soil pH is the main driver of archaeal community structure in soils. This strong correlation between soil pH and the microbial community structure may be because pH and the other physicochemical soil parameters are so closely related. Most microorganisms have intracellular pH that varies in the range of pH 7±1; therefore, a small variation of the environmental pH could expose microorganisms to stress. Archaea were detected in all analyzed samples and showed very low diversity compared to the bacterial community found in the same samples.18 Interestingly, Floresta decidual presented the lowest alpha diversity when compared to the other areas (Fig. 3). It has been suggested that an environment in equilibrium, such as forest, the ecosystem functioning is maintained based on lower levels of diversity, but high abundance of microorganisms; in contrast, environments under stress would present an increased diversity, which leads to a higher functional diversity and, consequently, the maintenance of essential ecosystem functions.30 Considering that Cerrado areas are more prone to environmental stress, such as high temperature, low moisture and oligotrophy, the higher archaeal diversity would be expected.
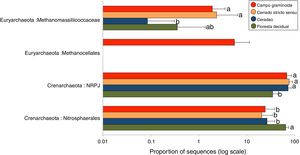
A total of 3078 reads were clustered in 33 OTUs according to the Greengenes taxonomical classification. We found Crenarchaeota to be the most abundant phyla across the Cerrado gradients, among which Nitrosphaerales and NRPJ were the predominant classes (Fig. 4). Organisms belonging to the largely non-thermophilic Crenarchaeota appear to be ubiquitous in soil systems and are dominant over euryarchaeal populations in grassland soils.31
Average relative abundance of the most abundant archaeal groups in soils from Campo graminoide, Cerrado stricto sensu, Cerradao and Floresta decidual as revealed by the 16S rRNA gene ribotyping. For each sample type, the number of replicates is n=9. Different lower letters indicate significant difference (FDR, P<0.05) among samples.
These findings agree with previous studies in the Brazilian Cerrado, which have reported Crenarcheota as most dominant phylum and Nitrosphaerales as an important group.3,16 The relative abundance of Nitrosphaerales and NRPJ differed across the regions. Floresta decidual showed the highest abundance of Nitrosphaerales, whereas Campo graminoide, Cerrado stricto sensu and Cerradao showed the highest abundance of NRPJ. Previously, Nicol et al.32 have indicated that Crenarchaeota from soils may have specific association with plant roots, playing an important role in the rhizosphere. Nitrosphaerales are important ammonia-oxidizer organisms,33 and although ammonia oxidation occurs in both Bacteria and Archaea, the archaeal group dominates in both soil and marine environments.34 Nitrogen-related microorganisms are commonly found abundantly in forest soils,30 which may explain the higher abundance of Nitrosphaerales in our forest samples. Also, Navarrete et al.35 showed that the diversity of ammonia-oxidizing archaeal communities tend to be higher in primary forests. On the other hand, the NRPJ group was more abundant in Cerrado sites. This group belongs to the Marine Benthic Group A, an uncultured archaeal cluster initially described from cold continental slope and deep-sea sediments.36 Members of this group are commonly found in oligotrophic soils, such as boreal forests,37 which may explain its higher abundance in our Cerrado samples. In summary, our results showed a high abundance of Crenarchaeota groups in our samples, with differential abundance between sites, suggesting that members of this group play distinct and important roles in the soil ecosystem.
The phylum Euryarchaeota is considered the most physiologically diverse and includes methanogens, halophiles and thermophile organisms. Members of this phylum were also found across the Cerrado gradients, including the classes Metanocellales and Methanomassiliicoccaceae. Interestingly, the Metanocellales class was detected only at Campo graminoide, which also had a high abundance of Methanomassiliicoccaceae. Similarly, Cerrado stricto sensu showed high abundance of Methanomassiliicoccaceae. The family Methanomassiliicoccaceae comprises of a methanogenic lineage of the class Thermoplasmata, and members of the Methanocellales family are methanogenic organisms, contributing to the global methane cycle.38 In this sense, our results show that important groups related to carbon cycle are abundant in Campo graminoide and Cerrado stricto sensu.
As shown in the RDA, pH was the main driver of the archaea communities across the gradients. It has been described that pH dominates the variation of archaeal diversity and community composition in tropical soils.39 In a large-scale study, the authors showed a niche specialization of terrestrial archaeal ammonia oxidizers based on pH, showing that amoA abundance and diversity increased with soil pH.40 In our samples, the high soil pH found in the Floresta decidual area favored the dominance of Nitrosphaerales, which is sensitive to low pH.41 On the other hand, a high abundance of Euryachaeota, specifically Metanocellales, in the Campo graminoide region may be related to the low soil pH found in this area. According to Hu et al.,41 soil pH is a driver of methanogenesis and subsequently may promote shifts in the archaeal community composition from Crenarchaeota to methanogenic Euryarchaeota.
In conclusion, our data showed that although the most of detected taxa are shared between all four areas, the archaeal structure and abundance differed across these gradients and was strongly driven by soil physicochemical properties. The soil pH was the main driver of archaeal communities in the studied soils, and this can be explained by the difference between the sites, where Floresta decidual presented higher values while Campo graminoide was more acid. Results showed that Crenarcheota was the most dominant phylum, followed by Euryarchaeota. Additionally, differential soil parameters lead to distinct functional potential of the communities, where Floresta decidual presented high abundance of groups related to the nitrogen metabolism while Campo graminoide and Cerrado stricto sensu presented high abundance of groups related to carbon metabolism. Also, Nitrosphaerales was the most abundant order found across the gradients, and further studies are needed to evaluate their potential in N cycling.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank “Fundação de Amparo a Pesquisa no Estado do Piauí” (FAPEPI) for financial support to this project through of PRONEX (FAPEPI/CNPq 004/2012). We thank the Unidade Multiusuário do Núcleo de Pesquisa e Desenvolvimento de Medicamentos (UM-NPDM) from Federal University of Ceará for using MiSeq Illumina sequencing facilities.