Although antibiotic-resistant pathogens pose a significant threat to human health, the environmental reservoirs of the resistance determinants are still poorly understood. This study reports the detection of resistance genes (ermB, mecA, mupA, qnrA, qnrB and tetL) to antibiotics among certain culturable and unculturable bacteria associated with the marine sponge Petromica citrina. The antimicrobial activities elicited by P. citrina and its associated bacteria are also described. The results indicate that the marine environment could play an important role in the development of antibiotic resistance and the dissemination of resistance genes among bacteria.
The spread of antibiotic-resistant microorganisms in the environment is globally recognized as an important public health issue, and there are concerns on our future ability to treat infectious diseases.1 Therefore, the knowledge of the nature of these resistance determinants in natural habitats is indispensable to get a better insight of the development of antibiotic resistance in clinical settings.2
In a previous publication, Marinho and colleagues3 demonstrated the antimicrobial and cytotoxic activities of the compound halistanol trisulphate isolated from P. citrina. This compound exhibited a broad-spectrum antibacterial activity against certain medically important bacteria, including resistant strains of Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Mycobacterium fortuitum and Neisseria gonorrhoeae.3
Symbiotic microbial communities can significantly impact the host-sponge ecology and evolution through supplemental nutrition and by the production of bioactive substances that can deter predators, competitors, and fouling organisms. Many of these substances possess antibacterial activity.4 The microbes that produce these antibiotics harbor resistance genes to protect themselves. Therefore, the selective pressure of the environment shapes these bacterial communities.5
In this background, the aim of the present study was to detect the resistance genes in culturable and unculturable bacteria associated with the sponge P. citrina. This study is the first report detecting the antibiotic resistance genes in P. citrina by culture-independent approaches. Such genes have usually been described in pathogenic bacteria.
Material and methodsSponge collection and bacteria used in this studyThe samples of the sponge P. citrina were collected by scuba-diving at a depth of 4–20m at Cagarras Archipelago (23801′S–43811′W), located in Rio de Janeiro, south-eastern Brazil (south-western Atlantic).
The bacterial strains were isolated and identified from P. citrina by Santos-Gandelman and colleagues in an earlier study.6 Of them, six were selected according to their antibacterial activity against certain medically important strains7 and/or antibiotic resistance profile.6Bacillus Pc31 and Pc32, Enterococcus Pc5b and Shigella Pc5a strains were grown in brain–heart infusion medium (BHI) (Difco, MI, USA), and Bacillus Pc3M and Halomonas Pc51M were grown in a marine medium (Marine 2216, Difco), at 25°C for 24h.
The following strains were included as positive controls for specific amplification of the different genes under investigation: Escherichia coli LO (qnrA), E. coli EB2b (qnrB), Streptococcus agalactiae (ermB), S. agalactiae CL5596 (tetL), Staphylococcus haemolyticus MD2 (mecA and mupA). These strains were grown in BHI medium at 37°C for 18h.
Polymerase chain reaction amplificationDNA from 0.25g of the sponge body was extracted using the Ultra Clean Soil DNA isolation kit (Mo Bio, Carlsbad, CA, USA) following the manufacturer's protocol. DNA from the bacterial strains was isolated by the guanidinium thiocyanate extraction method.8
Thus, the total DNA isolated from the bacteria from the sponge samples and from the culturable bacteria isolated from P. citrina were used to amplify genes conferring resistance to macrolide-lincosamide-streptogramin (ermB), methicillin (mecA), mupirocin (mupA), quinolones (qnrA, qnrB), and tetracyclines (tetL).
The following primers were used: for ermB, F: 5-CATTTAACGACGAAACTGGC and R: 5-GGAACATCTGTGGTATGGCG,9 to give a 425-bp product; for mecA, F: 5-TAGAAATGACTGAACGTCCG and R: 5-TTGCGATCAATGTTACCTAG,10 to give a 154-bp product; for mupA F: 5-GTTTATCTTCTGATGCTGAG and R: 5-CCCCAGTTACACCGATATAA,11 to give a 237-bp product; for qnrA, F: 5-ATTTCTCACGCCAGGATTTG and R: 5-GATCGGCAAAGGTTAGGTCA,12 to give a 516-bp product; for qnrB, F: 5-GATCGTGAAAGCCAGAAAGG and R: 5-ACGATGCCTGGTAGTTGTCC,12 to give a 469-bp product; for tetL, F: 5-ATAAATTGTTTCGGGTCGGTAAT and R: 5-AACCAGCCAACTAATGACAATGAT,13 to give a 1077-bp product.
The reaction mixtures, in final volumes of 50μL, contained MgCl2 (1.5mM for the mecA and mupA genes; 2mM for the ermB and tetL genes, and 4mM for the qnrA and qnrB genes), deoxynucleoside triphosphates (0.2mM each), primers (0.5μM each), Taq DNA polymerase (0.5U), reaction buffer (10mM), and 10–20ng of the extracted DNA as the template.
The PCR conditions were initial denaturation at 94°C for 5min, followed by 32 cycles at 94°C for 45s, 53°C for 45s, and 72°C for 60s, with a final elongation step at 72°C for 5min.12 The positive (strains with known resistance genes) and negative (without DNA template) controls were included in each run. Amplification products were provisionally identified from their sizes in ethidium bromide-stained agarose gels.
Results and discussionThe information about the selection pressures on antibiotic resistance genes is very limited regarding the remote environments with low direct human contacts. A more comprehensive understanding of the natural roles of putative antibiotic resistance genes is crucial in understanding of their origin and functions.14
In recent years, several antibiotics and other bioactive molecules have been isolated from marine sponges15 and from sponge-associated bacteria,4,16 including P. citrina3 and its associated bacteria.7
The P. citrina samples were collected at Cagarras Archipelago, which is a recent marine protected area located on the coast of Rio de Janeiro, Brazil. These islands are impacted both by the Guanabara Bay waters and by the discharges from a submarine outfall that releases untreated domestic sewage, both of which are balanced by the influx of pristine offshore water masses.17
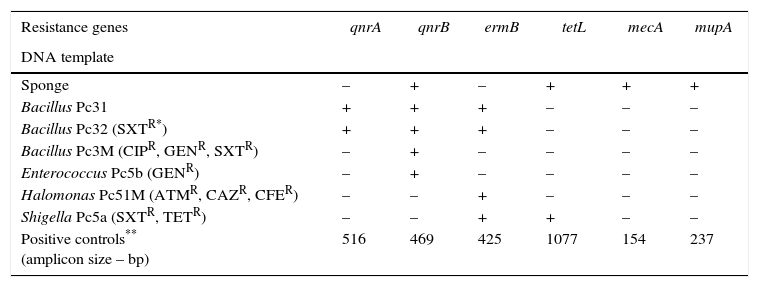
In this study, resistance genes for different antibiotics were detected in the DNA extracted from the culturable and unculturable bacteria associated with the sponge P. citrina. All amplicons were of the sizes of those of the positive controls (Table 1). The antibiotic resistance profile of the culturable bacteria associated with P. citrina has already been reported.6 This conforms to the data reported herein, as we have reported genes for quinolone and erythromycin resistance. Besides, the results also indicate that the hologenome of P. citrina contains genes encoding antibiotic resistance to erythromycin, methicillin, mupirocin, quinolone, and tetracycline. This goes in line with the fact that many marine sponges harbor dense and diverse microbial communities of considerable ecological and biotechnological importance.5
Antibiotic resistance genes detected in the culturable and unculturable bacteria associated with the sponge P. citrina.
| Resistance genes | qnrA | qnrB | ermB | tetL | mecA | mupA |
|---|---|---|---|---|---|---|
| DNA template | ||||||
| Sponge | – | + | – | + | + | + |
| Bacillus Pc31 | + | + | + | – | – | – |
| Bacillus Pc32 (SXTR*) | + | + | + | – | – | – |
| Bacillus Pc3M (CIPR, GENR, SXTR) | – | + | – | – | – | – |
| Enterococcus Pc5b (GENR) | – | + | – | – | – | – |
| Halomonas Pc51M (ATMR, CAZR, CFER) | – | – | + | – | – | – |
| Shigella Pc5a (SXTR, TETR) | – | – | + | + | – | – |
| Positive controls** (amplicon size – bp) | 516 | 469 | 425 | 1077 | 154 | 237 |
Antibiotic resistance to: aztreonam (ATMR), ceftazidime (CAZR), cephalexin (CFER), ciprofloxacin (CIPR), gentamicin (GENR), trimethoprim/sulfamethoxazole (SXTR), tetracycline (TETR) (Santos-Gandelman et al.6).
The application of culture-independent approaches, such as PCR and metagenomics, for the study of antibiotic resistance genes in the environment has uncovered a vast diversity of antibiotic resistance genes in soil bacteria. However, according to the best of our knowledge, this is the first time that mecA, mupA, qnrB and tetL were detected in sponge-associated bacteria by culture-independent approaches. These results demonstrate that PCR is also a powerful tool to detect potential antibiotic resistance genes in marine environments.
While many antibiotic-resistance genes are believed to have their origin in natural ecosystems, their abundance, nature, and ecological role in such settings remain relatively obscure. In addition, antibiotics used in therapeutics and agriculture are known to accumulate in the environment and to contaminate aquatic habitats, where they can exert a selective pressure on the native flora.18 Erythromycin, quinolone and tetracycline-resistant bacteria can be found even in pristine environments and in animals.19 Recently, some of these resistance genes have been identified in a Bacillus sp. isolated from the sponge Haliclona simulans.20 The plasmid-mediated quinolone resistance genes, qnrA and qnrB, have already been detected in bacterial strains isolated from aquatic environments.21 These genes are horizontally transferable among bacteria.18
Mupirocin, which is also known as pseudomonic acid A, is produced by Pseudomonas fluorescens isolated from soil environments.21 Plasmids that confer high-level resistance to mupirocin were isolated nearly twenty years before the clinical use of this drug.22 The mupA gene was also found in the bacterium, Oceanobacillus iheyensis, isolated from deep-sea sediments at a depth of over 1000m.23
The mecA gene is usually acquired along with a variety of genetic elements. The origin of mecA and the other genes of these cassettes have been the subject of an intensive research since the original discovery of methicillin-resistant Staphylococcus aureus (MRSA).24 Earlier works suggested homology with mec genes found in the coagulase-negative Staphylococcus sciuri group, which has been isolated from animals and food products, and occasionally from humans.25,26 Other authors speculated that mecA originated from Staphylococcus fleurettii, a species isolated from raw-milk cheese.27 While it has been reported that some antibiotic resistance genes might have originated from marine bacteria, it is probably not the case of mecA, as the relationship of Staphylococcus with the marine environment remains elusive.28
ConclusionsIt is important to characterize the resistance genes in the entire marine community, including both culturable and unculturable strains.18 Little is known about the antibiotic resistome of the vast majority of the environmental bacteria, although there have been calls for a better understanding of the environmental reservoirs of antibiotic resistance and their potential impacts on clinically important bacteria.29 The prevalence and diversity of the resistance genes in the environment inspire hypotheses about the native roles of these resistance genes in the natural microbial communities. Considering that antibiotic treatment is our primary, and in many cases only, method of treating infectious diseases, we conclude that more detail studies of the environmental reservoirs of the resistance genes are crucial for our ability to fight infections in future.
FundingThis work was supported by grants from CAPES, CNPq and FAPERJ to M.S. Laport. D.S. Santos, J.F. Santos-Gandelman and P.V.M. Pontes received FAPERJ, CAPES and CNPq fellowships, respectively.
Conflict of interestThe authors declare no conflict of interest.
The authors give special thanks to Dr. Walter Oelemann for his assistance in the preparation of this manuscript and to Dr. Kátia Regina Netto dos Santos, Dr. Lucia Martins Teixeira and Dr. Renata Picão for providing bacterial strains used as the controls in the PCR reactions.