To identify genes involved in the pathogenic mechanisms of non-proliferative diabetic retinopathy (NPDR), among which include oxidative stress, extracellular matrix changes, and/or apoptosis, in order to evaluate the risk of developing this retinal disease in a type 2 diabetic (DM2) population.
Material and methodsA case–control study was carried out on 81 participants from the Valencia Study on Diabetic Retinopathy (VSDR) of both genders, with ages 25–85 years. They were classified into: (i) DM2 group (n=49), with DR (+DR; n=14) and without DR (−DR; n=35), and (ii) control group (GC; n=32). The protocols included a personal interview, standardized ophthalmological examination, and blood collection (to analyze the DNA for determining the gene expression (TP53, MMP9, and SLC23A2) in the study groups). Statistical analyses were performed using the SPSS v22.0 program.
ResultsThe TP53 and MMP9 genes showed a higher expression in the DM2 group compared to the GC, although the difference was only significant for the MMP9 gene (TP53: 10.40±1.20 vs. 8.23±1.36, p=0.084; MMP9: 1.45±0.16 vs. 0.95±0.16, p=0.036), and the SLC23A2 gene showed a significant lower expression in the DM2 vs. CG (5.58±0.64 vs. 11.66±1.90, p=0.026). When sub-dividing the DM2 group according to the presence of retinopathy, the expression of the TP53, MMP9 and SLC23A2 genes showed significant differences between the DM2−RD, DM2+RD and GC groups (TP53: 9.95±1.47 vs. 11.52±2.05 vs. 8.23±1.36, p=0.038; MMP9: 1.47±0.20 vs. 1.41±0.27 vs. 0.95±0.16, p=0.021; SLC23A2: 5.61±0.77 vs. 5.51±1.21 vs. 11.66±1.90, p=0.018).
ConclusionsGenes involved in extracellular matrix integrity (MMP9) and/or apoptosis (TP53), could be considered potential markers of susceptibility to the development/progression of NPDR. Interestingly, the SLC232A2 gene (ascorbic acid transporter) can be considered a protector of the risk of the development/progression of the retinopathy.
Identificar genes implicados en los mecanismos patogénicos de la retinopatía diabética no proliferante, como estrés oxidativo, alteración de la matriz extracelular y/o apoptosis, para valorar el riesgo de desarrollo de la misma en una población de diabéticos tipo 2 (DM2).
Material y métodosEstudio de casos y controles en 81 participantes del Estudio Valencia sobre Retinopatía Diabética (EVRD), de ambos sexos y con edades comprendidas entre los 25 y 85 años, clasificados en: 1) grupo DM2 (n = 49), con RD (+RD; n = 14) y sin RD (−RD; n = 35), y 2) grupo control (GC; n = 32). Se realizó entrevista personal, examen oftalmológico estandarizado y extracción de sangre que se procesó para analizar el ADN y determinar la expresión de: TP53, MMP9 y SLC23A2 en todos los participantes. El programa estadístico utilizado fue el SPSS v22.0.
ResultadosLos genes TP53 y MMP9 aumentaron su expresión en el grupo DM2 respecto al GC, aunque solo de manera significativa el gen MMP9 (TP53: 10,40 ± 1,20 vs. 8,23 ± 1,36, p = 0,084; MMP9: 1,45 ± 0,16 vs. 0,95 ± 0,16, p = 0,036) y el gen SLC23A2 disminuyó significativamente sus niveles en DM2 vs. GC (5,58 ± 0,64 vs. 11,66 ± 1,90, p = 0,026). Al subdividir el grupo DM2 según presencia de retinopatía, la expresión de los genes TP53, MMP9 y SLC23A2 mostró diferencias significativas entre los grupos DM2−RD, DM2+RD y GC (TP53: 9,95 ± 1,47 vs. 11,52 ± 2,05 vs. 8,23 ± 1,36, p = 0,038; MMP9: 1,47 ± 0,20 vs. 1,41 ± 0,27 vs. 0,95 ± 0,16, p = 0,021; SLC23A2: 5,61 ± 0,77 vs. 5,51 ± 1,21 vs. 11,66 ± 1,90, p = 0,018).
ConclusionesLos genes reguladores de apoptosis (TP53) e integridad de la matriz extracelular (MMP9) podrían estar implicados en la susceptibilidad para el desarrollo/progresión de la RD, así como el gen SLC232A2 (transportador del ácido ascórbico) puede comportarse como protector del riesgo de padecer/progresar en la retinopatía.
Diabetes mellitus (DM) is continuously increasing throughout the world. This prevalence is due mainly to type 2 (DM2), related to obesity and sedentary lifestyles, which has become a large magnitude socio-sanitary problem both in adults1 and children.2 This calls for early detection programs and the development of validated biomarkers allowing for including molecular and genetic studies in the clinic in order to prevent the presentation of DM and the complications derived from the chronic nature of the process, including diabetic retinopathy DR.1–3
The development of DM2 requires the coexistence of endogenous and exogenous factors, some of which are potentially modifiable such as overweight and the lack of physical exercise that produce insulin secretion alterations in response to hyperglycemia stimulation as well as changes in hormonal action on peripheral tissue. However, epidemiological and experimental studies have demonstrated that, in the context of hyperglycemia, arterial hypertension and dyslipidemia are significant factors but not sufficient to develop chronic DM2 complications.4
Even though the etiopathogeny of DM2 involves several mechanisms such as inflammation, angiogenesis and apoptosis among others, the genetic factor is crucial to complete the knowledge of the cellular and molecular background of the disease.5–7 Even so, all the genes involved in the development of DM are not known, not even those related with the clinic presentation form and the pharmacological response.
DR is the main cause of visual loss throughout the world in the working age population. Approximately one third of the nearly 300 million diabetics exhibit signs of DR, and of these nearly a third exhibit severe complications in the course of the disease, including retina detachment, neovascular glaucoma or diabetic macular edema.8–10 Recent studies have described numerous biological pathways involved in DR pathogeny including inflammation, oxidative/nitrosative stress, the alteration of some hormones related to metabolic pathways (leptin and adiponectin), vitamins C and D and apoptosis.8–16
It has been suggested that progression of DR is genetically determined, mainly the evolution from nonproliferative to proliferative forms of DR.10,16,17 However, not all DM2 patients evolve to DR nor do they evolve in a similar manner; for instance diabetics who exhibit advanced forms of retinopathy with a very short DM2 evolution. In addition, individual response to medicaments is also highly variable. In some cases, drugs could delay or halt the course of retinopathy although the process has not been reversed in any patient.10,15,16 Even though there is increasing evidence that genetic alterations contribute to the development of DR, the identification of genes is very difficult. For this reason, a range of techniques have been developed, some of which are briefly described below.18–20
It is possible that the most direct way to identify a candidate gene to susceptibility for the disease is the identification of a chromosome alteration (that alters one or more genes, i.e., cytogenetic abnormality). However, it must be taken into account that cytogenetic alterations and ligation analyses are strategies for identifying genes that account for diseases in familial cases, and that association studies are adequate for sporadic cases. A further possibility is the ligation and positional clonation analysis (localization of the culprit gene on the basis of its chromosome position, narrowing an interval that contains the diseased gene in a specific chromosome). On the other hand, association analysis is based on the fact that 2 alleles ligated in the same chromosome will continue to transmit together in each meiosis until separated by meiotic recombination, at which point they will distribute in different gametes and establish independent associations. Association studies endeavor to find genetic marker correlations for the disease through testing ligation imbalance in a specific population, acquiring greater potency to detect alleles for risk of slight/moderate disease when compared to ligation analysis. This technique comprises several variants such as the association study of positional candidate genes (in which direct association gene tests are applied following the ligation analysis that previously identified a chromosome region, identifying genes in that region and limiting the number of genes to be screened), and the functional association studies of candidate genes (an alternative to the previous one which does not require knowing the chromosomal location of the gene).
One of the most studied genes in relation to DR is the vascular endothelial growth factor (VEGF), which gives rise to typical proliferative DR neovascularization and the appearance and development of diabetic macular edema. Due to its clear relationship with the etiopathogenic mechanisms of the disease, multiple VEGF gene polymorphisms have been studied and new data on variants of this gene possibly associated to DR are published every day. Even though in many cases said association is weak or inconsistent, others provide sufficient evidence of the involvement of VEGF gene variants in the risk of expressing DR.21–23 In addition to said gene, many others have been associated to retinopathy, with new studies being published frequently. Genes such as SLC2A11, SLC24A3, DDR2, HTR1B, INSR, TNFα or IL10 are associated to the risk of DR24,25 whereas others like eNOS3 seem to play a protective role.26
Despite all the efforts, the researchers who endeavored to identify genes exhibiting genetic susceptibility to DR or progression for the disease were unable to contribute breakthroughs in the past 10 years due to contradictory results, different study populations and the lack of data reproducibility.27–35 In addition, many genes which have not yet been studied, such as SLC23A2, MMP9 and TP53, could still be potentially involved in the development and/or progression of DR.
Solute Carrier family 23 member 2 [SLC23A2] is a protein coded in humans by the SLC23A236 gene whose function is to transport ascorbic acid (vitamin C). There are notable differences between the consumption of vitamin C and its levels in peripheral blood and tissue. This sometimes depends on modifiable factors such as tobacco smoking, obesity and hyperglycemia. In fact, a negative correlation has been suggested between the daily intake of vitamin C and the risk of DM2.37
Extracellular matrix proteases such as the metalloproteinases (MMP) family are involved in the rupture of the matrix in normal and pathological processes. Some MMPs are secreted as inactive pro-proteins that become active when joining extracellular proteinases. The enzyme that codes for the MMP9 gene degrades collagen type IV and V, as well as other matrix proteins under normal and pathological situations.38 Recently, altered MMP9 levels have been described in patients with DM2, as well as an association between the MMP9-1562C/T polymorphism and diabetic complications.39
P53 tumor proteins, also known as Transforming Related Protein 53 [TRP53], are coded in humans by the TP5340 tumor suppression gene that regulates genomic stability (cell cycle, senescence, apoptosis) and prevents mutations, for which reason it is known as “the genome guardian”. At present, at least 12 proteins are known to be coded by said gene. It has recently been described to be involved in DM.41 At the ocular level, an experimental study of the authors’ group has demonstrated its participation in the regulation of endogenous stress in the retina and optic nerve of transgenic rats.42
The authors have carried out a study designed to identify genes related to oxidative stress (transport of antioxidant vitamins) such as SLC23A2, extracellular matrix integrity such as MMP9 and apoptosis regulation such as TP53, in a group of patients with DM2 with and without retinopathy, to find different expressions and comparing them with a group of healthy subjects in order to establish their involvement in the onset and progression of DR.
MethodA case and control study paired by age and gender, selected from the Valencia Study on Diabetic Retinopathy (VSDR)15 database. The present study corresponds to Report number 3 in accordance with the inclusion/exclusion criteria proposed for said study (Table 1), with the purpose of identifying genes related to the pathogenic mechanisms of retinopathy in patients with DM2. The study was approved by the ethics and clinic research committees of the appropriate centers. All the experiments were adjusted to the standards on human research (Helsinki Declaration on Human Experimentation; Helsinki 1964, updated 2004 version) and in accordance with the current regulations of the European Community for genetic studies. The participants were informed and signed consents to participate in the study.
Inclusion/exclusion criteria for the study.
| Group of DM2 patients | Control Group |
|---|---|
| Inclusion | |
| Age 25–85 years | Age 25–85 years |
| Diagnostic of DM2 for at least 5 years. Patients with or without retinopathy | No DM (types 1 or 2). No retinopathy |
| No chronic or degenerative systemic disease | No chronic degenerative systemic disease |
| Exclusion | |
| Age <25 years or >85 years | Age <25 years or >85 years |
| Diagnostic of DM2<4 years | Having DM (types 1 or 2) |
| Having chronic or degenerative systemic disease | Having chronic or degenerative systemic disease |
DM: diabetes mellitus; DM2: diabetes mellitus type 2.
The sample was calculated by means of the eNe 2.0 application (Glaxo Smith Kline S.A.), obtaining a statistical power of 80% and detecting differences in the comparison of working hypotheses (Ho: p1=p2) and utilizing the χ2 bilateral test for 2 independent samples, considering a significance level of 5%. Accordingly, the sample of participants comprised 81 individuals of both sexes and between 25 and 85 years of age, who were classified as: (1) group DM2 (n=49), with DR (+DR; n=14) and without DR (−DR; n=35), and (2) control group of non-diabetic subjects without retinopathy (GC; n=32).
Operational definitionA personal interview was carried out that included data on DM2 (onset, duration, treatment and family history), personal characteristics (other diseases and treatments, height and weight) and lifestyle (nutrition, physical exercise smoking/drinking habit, etc.).
A standardized ophthalmological examination was carried out comprising best corrected visual acuity (BCVA), intraocular pressure (IOP), anterior and middle segment biomicroscopy, ocular fundus study with dilatation and examination with 78D lens, retinographies (TOPCON ImageNet TRC-50JA, Barcelona, Spain) retina nerve fiber layer central thickness measured with spectral domain optical coherence tomography (SD-OCT) (Carl Zeiss Meditec, Madrid, Spain). The presence or absence of DR was based on funduscopic findings and descriptions of the updated Retina and Vitreous Society of Spain regulations.43 Retinopathy was classified in accordance with the international DR severity scale of the Early Treatment Diabetic Retinopathy Study [ETDRS]44 and the current regulations of the American Academy of Ophthalmology.45
In order to obtain biological samples, preprandial blood extractions were scheduled for analyzing gene expression that was performed at the lab of the Ophthalmological Research Unit «Santiago Grisolía»/FISABIO, Cellular and Molecular Ophthalmobiology Unit of Valencia University and at the Preventive Medicine and Public Health Dept. of Valencia University. The procedure is briefly explained below.
Extraction and quantification of RNA in bloodFirst, serum was separated through sedimentation and centrifuged at 2000 RPM during 15min. After removing the supernatant, the pellets were washed several times with PBS, subsequently adding trizol (500–1000μl, depending on pellet size) and storeing at −80°C until the following day. After defreezing, the samples were homogenized to ensure steady rupture. Chloroform was added and the samples were centrifuged (13,000rpm, 15min). The aqueous phase was transferred to a new tube, isopropanol was added and the samples were left at −20°C through the night. The following day they were centrifuged (13,000rpm, 15min) removing the supernatant. 75% cold ethanol was added and centrifuged again (13,000rpm, 15min, at 4°C). Finally, after removing ethanol completely, the pellets were suspended again in 20μl of water free of RNases, the RNA was quantified with spectrophotometry (NanoPhotometer, Implen GmbH, Schatzbogen 52, D-81829, Germany) and the samples were stored at −80°C.
Conversion to complementary DNA (cDNA) and quantificationRNA was converted to cDNA utilizing the High Capacity RNA-to-cDNA kit (Ref. 4387406. Applied Biosystems, Foster City, CA, USA), following manufacturer instructions. 300ng of RNA of each processed blood sample of each participant were retrotranscribed. PCR conditions were: 5min/25°C, 30min/42°C and 5min/85°C. quantification was carried out with spectrophotometry (NanoPhotometer, Implen GmbH, Schatzbogen 52, D-81829, Germany).
Expression analysis of studies genesThe relative expression of TP53, MMP9 and SLC23A2 genes was analyzed with real-time qPCR, utilizing Taqman probes and 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The samples were analyzed twice, using the GAPDH gene (3-glyceraldehyde phosphate dehydrogenase) as the reference (housekeeping) gene. PCR conditions were: 2min/50°C, 10min/95°C, and 40 cycles 15s/95°C and 1min/60°C.
Statistical analysisThe statistical application utilized for analyzing all data was SPSS 22.0. (IBM SPSS Statistics for Windows 22.0 program, IBM Corp., Armonk, NY, USA). Normality for quantitative variables was determined with the Kolmogorov–Smirnov test. Categorical variables were compared utilizing Pearson's χ2 test. Mean comparison was carried out with the Mann Whitney U test for non-abnormal variables. Comparisons of more than 2 mean values was done with the Kruskal Wallis test. Spearman's Rho was utilized to determine the magnitude of association of 2 quantitative variables.
The relative expression of each gene was calculated utilizing the double ΔCt formula.46
All the statistical analyses were made assigning a significance value of 0.05.
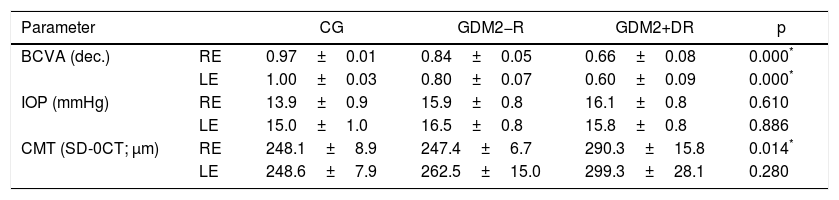
ResultsEye fundus findings regarding DR diagnostic were: DM2+DR, n=14 (28.6%) and DM2−DR, n=35 (71.4%). Table 2 shows the ophthalmological examination values obtained from study participants.
Comparison of ophthalmological parameters in groups of patients with DM2 (with/without DR) and healthy controls.
| Parameter | CG | GDM2−R | GDM2+DR | p | |
|---|---|---|---|---|---|
| BCVA (dec.) | RE | 0.97±0.01 | 0.84±0.05 | 0.66±0.08 | 0.000* |
| LE | 1.00±0.03 | 0.80±0.07 | 0.60±0.09 | 0.000* | |
| IOP (mmHg) | RE | 13.9±0.9 | 15.9±0.8 | 16.1±0.8 | 0.610 |
| LE | 15.0±1.0 | 16.5±0.8 | 15.8±0.8 | 0.886 | |
| CMT (SD-0CT; μm) | RE | 248.1±8.9 | 247.4±6.7 | 290.3±15.8 | 0.014* |
| LE | 248.6±7.9 | 262.5±15.0 | 299.3±28.1 | 0.280 |
BCVA: best corrected visual acuity; CMT: central macular thickness; CG: control group; GDM2+DR: group with diabetes mellitus type 2 and diabetic retinopathy; GDM2−DR: group with diabetes mellitus type 2 without diabetic retinopathy; RE: right eye; LE: left eye; IOP: intraocular pressure.
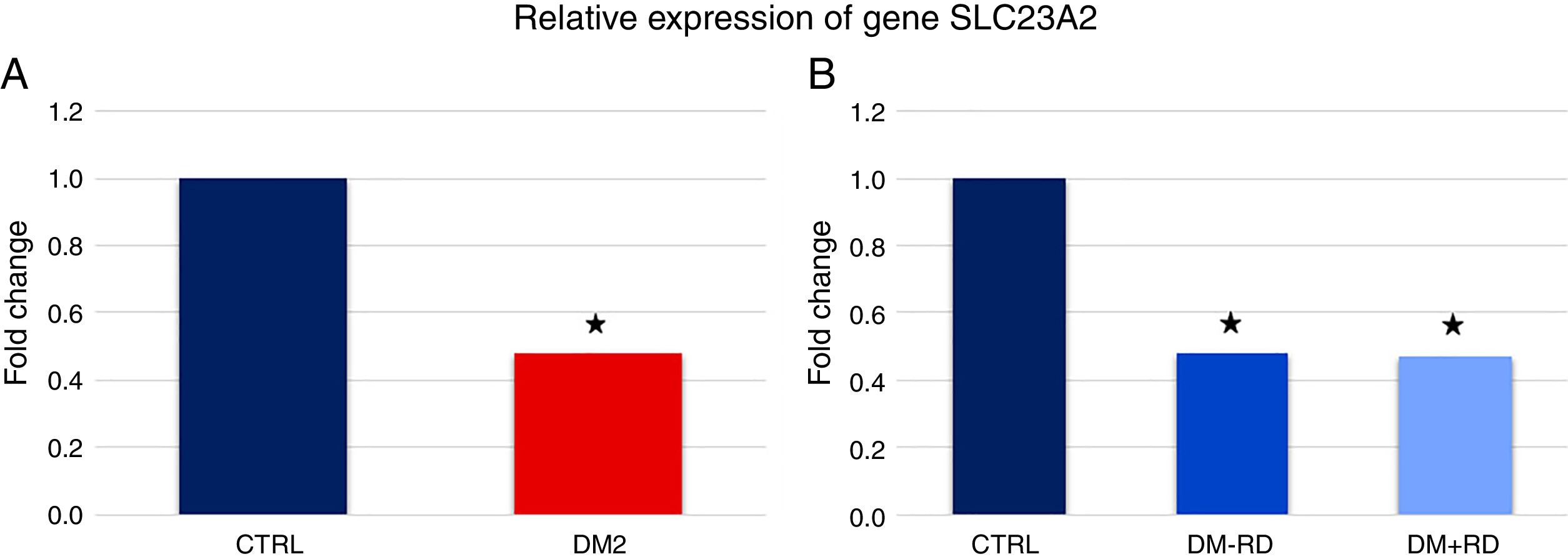
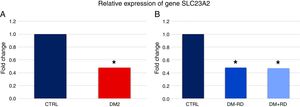
Fig. 1A shows that the expression of gene SLC23A2 was significantly lower in DM2 vs. GC (5.58±0.64 vs. 11.66±1.90, p=0.026). Subdividing the group of diabetic patients on the basis of presence of DR and comparing the expression of the SLC23A2 gene between the 3 groups, significance vis-à-vis the control group was maintained (DM2−DR: 5.61±0.77, DM2+DR: 5.51±1.21, GC: 11.66±1.90; p=0.018) (Fig. 1B).
Relative expression of gene SLC23A2 in study participants. (A) Comparison between the group of type 2 (DM2) diabetics and the control group (CTRL). (B) Comparison between the group of diabetics type 2 without diabetic retinopathy (DM2−DR), the group of diabetics type 2 with diabetic retinopathy (DM2+DR) and the control group (CTRL). The asterisk indicates statistically significant differences vis-à-vis the control group (p<0.05).
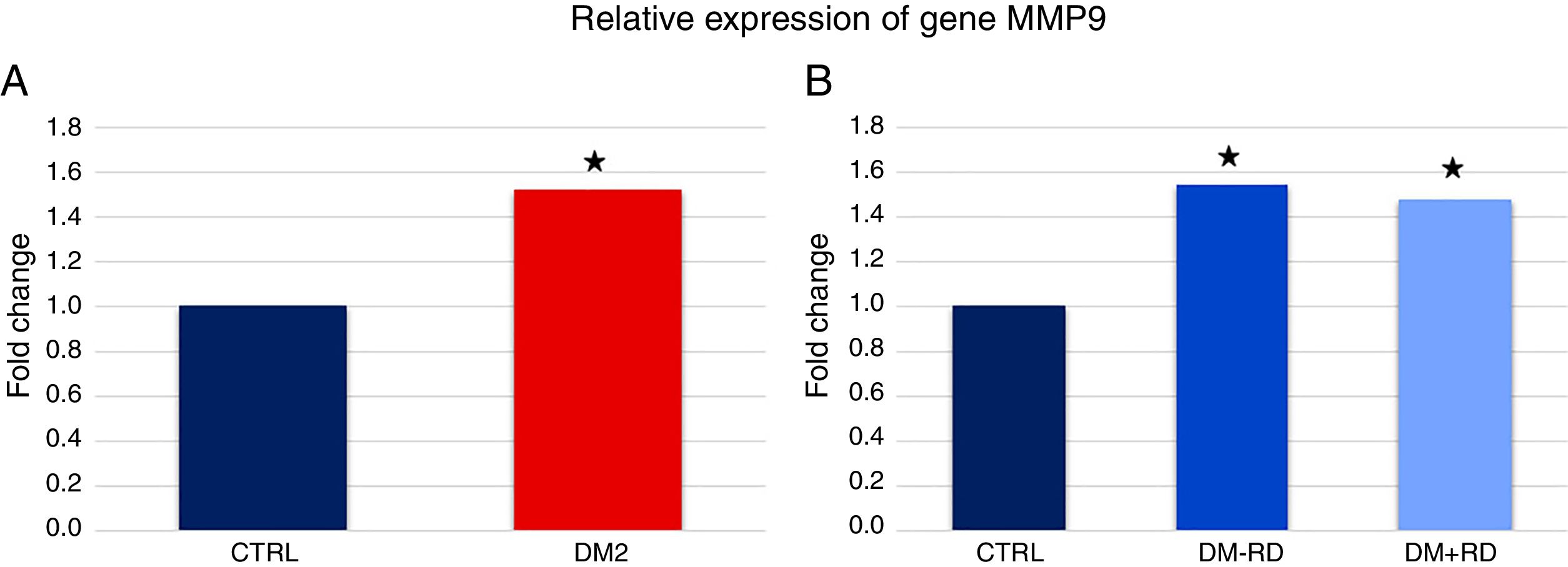
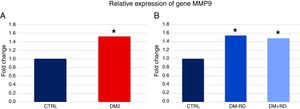
In what concerns gene MMP9, expression was significantly higher in DM2 vs. GC (1.45±0.16 vs. 0.95±0.16, p=0.036) (Fig. 2A). When comparing the 3 groups, the differences in the expression of MMP9 were also statistically significant (DM2−DR: 1.47±0.20, DM2+DR: 1.41±0.27, GC: 0.95±0.16; p=0.021) (Fig. 2B).
Relative expression of gene MMP9 in study participants. (A) Comparison between the group of type 2 (DM2) diabetics and the control group (CTRL). (B) Comparison between the group of type 2 diabetics without diabetic retinopathy (DM2−DR), the group of diabetics type 2 with diabetic retinopathy (DM2+DR) and the control group (CTRL). The asterisk indicates statistically significant differences vis-à-vis the control group (p<0.05).
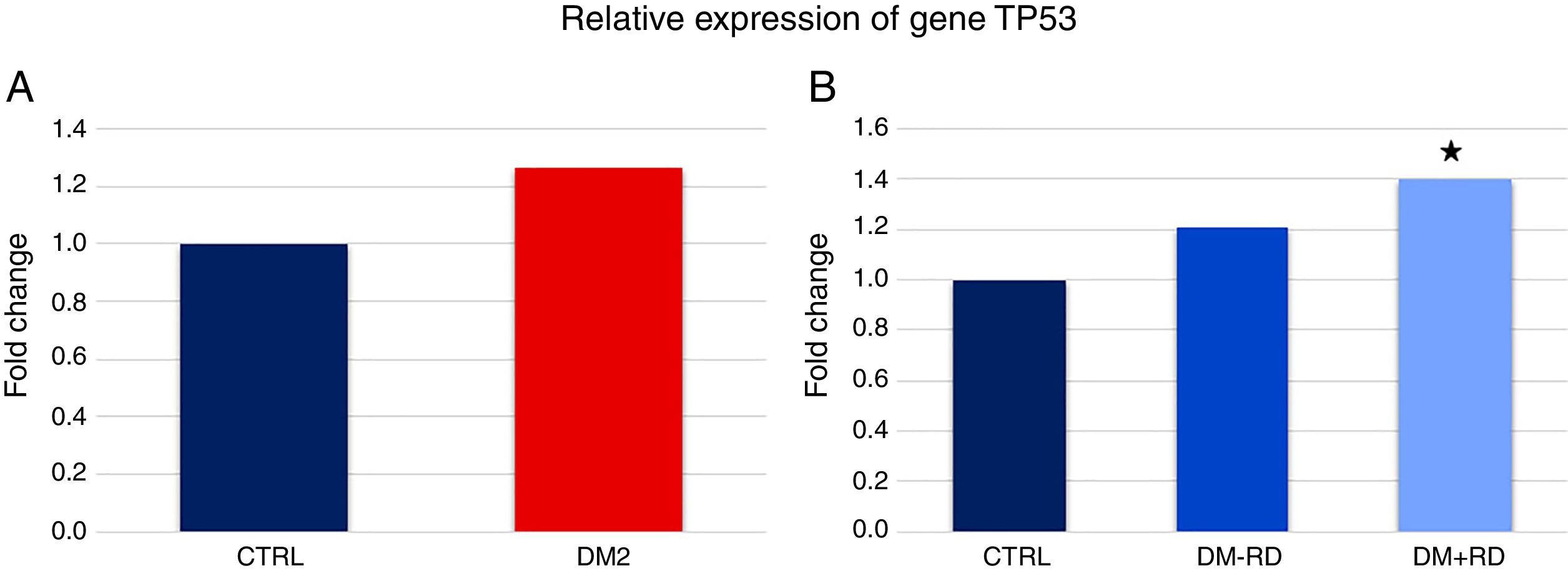
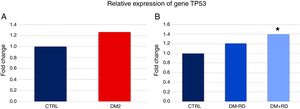
Lastly, the expression of TP53 was higher in DM2 vs. GC, although statistical significance was not reached (10.40±1.20 vs. 8.23±1.36, p=0.084) (Fig. 3A). However, a comparison between the 3 groups did show statistically significant differences (DM2−DR: 9.95±1.47, DM2+DR: 11.52±2.05, GC: 8.23±1.36; p=0.038) (Fig. 3B).
Relative expression of gene TP53 in study participants. (A) Comparison between el group of diabetics type 2 (DM2) and the control group (CTRL). (B) Comparison between the group of type 2 diabetics without diabetic retinopathy (DM2−DR), the group of diabetics type 2 with diabetic retinopathy (DM2+DR) and the control group (CTRL). The asterisk indicates statistically significant differences vis-à-vis the control group (p<0.05).
In none of the cases the expression of genes showed significant differences between the group of diabetics without retinopathy and the group with retinopathy.
DiscussionDR is the main cause of vision loss throughout the world. The prevalence of DR in Europe ranges between 22% and 37%, with a significant socio-sanitary cost. It is necessary to research the molecular mechanisms that underlie hyperglycemia, due to the risk of giving rise to DR as well as for the processes involved in the progression thereof and susceptibility to complications. Accordingly, this study has analyzed the expression of genes related to oxidative stress (transport of antioxidant vitamins), specifically gene SLC23A2, extracellular matrix integrity such as MMP9 and the regulation of apoptosis, specifically TP53, in a group of patients with DM2 selected from the VSDR database and divided in diabetics with and without DR, compared with a group of control subjects. The study data revealed a significant difference in the expression of the 3 genes between the DM2 and GC groups (p<0.05). In addition, gene SLC23A2 exhibited significant variations in its levels of expression in the presence or absence of retinopathy (p<0.05).
The mechanism that activates the cascade of cellular and molecular events that initiate the development of diabetic microangiopathy is precisely the alteration of the capillary base membrane and the loss of pericytes.3,10–17 It is known that, even at the pre-clinic level, it is possible to detect vascular dysfunction in retinal capillaries, involving a range of endothelial and smooth muscle processes. In addition, said early changes are the consequence of increased endothelines and diminished nitric oxide in the context of oxidative/nitrosative stress. These initial events continue with the increase of capillary permeability, involving processes such as extracellular matrix anomalies, ischemia-induced loss of pericytes and endothelial cells, and oxidative stress worsening. Vascular dysfunction progresses and presents capillary basal membrane thickening and the appearance of the remaining signs and symptoms that evidence the development of DR. Said process involves a countless number of signaling pathways and molecules.
It has been demonstrated that diminished vitamin C is a risk factor for developing DM. Vitamin C is a powerful antioxidant and cofactor of multiple enzymatic reactions, including collagen synthesis in connective tissue (skin, cartilage, teeth, bones and blood vessels). As the human body cannot synthesize vitamin C, it must be ingested in order to reach daily recommended values. Observational data of a study carried out in the United States, The National Health and Nutrition Examination Survey (NHANES I [1971–1974]; II [1976–1980] and III [1998–1994]) identified individuals with a recent DM diagnostic and saw they had low levels of vitamin C in serum.47–49 These findings were confirmed by other diabetic population studies50,51 that described the inverse correlation between the levels of vitamin C and HbA1c. The proteins that transport solutes such as vitamins are coded by the SLC gene. Solute Carrier family 23 member 2 [SLC23A2] is a protein coded in humans by the SLC23A2 gene,36 the main function of which is to transport vitamin C. the present study has observed that gene SLC23A2 was underexpressed in the diabetic subgroup, as shown in Fig. 1A. Similarly, the expression of this gene in diabetics with and without retinopathy was significantly below that observed in the control group (see Fig. 1B). Accordingly, it can be deduced that vitamin C levels are highly regulated by SLC23A2, which regulates vitamin C bioavailability and use by the organism. However, there are no studies relating the expression of said gene and DR. In what concerns other ocular processes, associations have been found between SLC23A2 variations (specifically, the rs1279683 polymorphism), vitamin C plasma concentration and the risk of glaucoma.52,53 The authors suggest that low expression of the SLC23A2 gene in patients with DR of this study involves a significant alteration of vitamin C transport that prevents its arrival at the target organs such as the retina where it will be unable to perform its antioxidant protective action at the time when the process is facilitating the development and/or progression of DR, because the increase of oxidative stress induces the activation of redox-sensitive transcription factors, thus changing the expression of many genes.
The extracellular matrix comprises a series of proteins, including collagen, elastine and others, proteoglycans and glycoproteins that confer structural properties to cells and tissue. Alterations in said matrix involve the loss of its functions which include nutrition, filtering, elimination, loss of regeneration and cicatrization capacities and mechanical transmission alterations, as well as the loss of the substrate for immune response. Extracellular matrix MMPs are in charge of degrading said matrix and for this reasons are involved in a range of normal and pathological processes, including DR.54 In fact, the MMP9 protein is increased in active neovascularization and has been demonstrated to have pro-apoptotic functions. The literature describes a mechanism by means of which DM activates MMP9 and its signaling chain with the participation of H-Ras, among others.55,56 In fact, hyperglycemia activates H-Ras, a low molecular weight G-protein, in the retina and its capillaries, the main effect of which is to provoke cell apoptosis in said vessels, pericytes and endothelial cells. The MMP9 protein is regulated by H-Ras and, in diabetes, its activation is associated with increased vascular permeability.57 Hyperglycemia activates said MMP-9 protein and the latter accelerates the apoptosis of retinal capillary cells, which in turn activate the development of DR.
The results of the present study match the above descriptions and maintain that the gene that encodes for the MMP9 protein is overexpressed in patients with DM2 vis-à-vis GC subjects, although it was not possible to demonstrate that their expression changes when having DR or not. However, other authors have described the association between the MMP9-1562C/T polymorphism and DM2 complications.39
As described above, apoptosis of retinal microcirculation cells begins the development of DR. The TP53 gene regulates cell cycle, senescence and apoptosis.58 No polymorphisms have been found in this gene in association with DR. However, there is evidence on the association of the Arg72Pro polymorphism with DM1 as well as DM2.59,60 The present study has found a significant increase of TP53 expression in patients with DM2 compared to GC, as suggested by other authors.61 In addition, other molecules such as cytokines, growth factors, hormones and oxidative stress metabolic products could over-regulate the expression of pro-apoptotic genes in the microvasculature, enhancing oxidative stress and ischemia to activate apoptosis and DR, as suggested previously for the retinal astroglia.62
In conclusion, the present study has identified genes involved in the transport of antioxidant vitamins, extracellular matrix homeostasis and apoptosis regulation. This study suggests that the deregulation of SLC23A2, MMP9 and TP53 promoted by chronic hyperglycemia is related to susceptibility for the Onset of DR and its progression toward the proliferative form.
Full list of VSDR researchersV. Zanón-Moreno, S.M. Sanz-González, J.J. García-Medina, A. LLeó-Perez, M.J. Roig-Revert, J. Marín-Montiel, V. Chaqués Alepuz, L. Alonso Muñoz, K. Shoaie Nia, L. Duarte, C. Campos Borges, O. Alvarez-Barrachina, J. Raga-Cervera, V. Chiner, M. Perez-Ramos, G. Roglá Navarro, M. Albert-Fort, M.F. García-Esparza, M. Saleh Rahhal, V. Vila-Bou, M. Villalonga, M. L. Redón, A. Llorca-Cardeñosa, R. Gallego-Pinazo, R. Dolz-Marco, M.I. López-Gálvez, D. Galarreta-Mira, L. Manzanas, E. Bendala-Tufanisco, F. Santander-Trentini, M. Salgado-Borges, A.L. López-Ramos, G. Virgili, C. Nucci, J.F. Arévalo, and Maria D. Pinazo-Durán.
FundingThis study has been partially funded by the FIS/FEDER PI13/00480 and PI16/00797 projects (IP: Dr. Pinazo-Durán) and with the Thea Laboratories 2013–2016 project (Barcelona, Clermond Ferrand; IP: Dra. Pinazo-Durán).
Conflict of interestNo conflict of interests has been reported by the authors in relation to this study.
Please cite this article as: Pinazo-Durán MD, Shoaie-Nia K, Sanz-González SM, Raga-Cervera J, García-Medina JJ, López-Gálvez MI, et al. Identificación de nuevos genes candidatos para la retinopatía en diabéticos tipo 2. Estudio Valencia sobre Retinopatía Diabética (EVRD). Informe n.° 3. Arch Soc Esp Oftalmol. 2018;93:211–219.