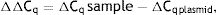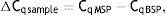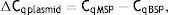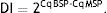Systemic lupus erythematosus (SLE) is an autoimmune disease caused by interaction of genetic, epigenetic, and environmental factors. One of the important epigenetic factors in SLE would be methylation of immune-related genes, such as FOXP3, which plays a role in activating the regulation and also the function of T cells. To date, the relationship between levels of serum bio-markers and the susceptibility to lupus in children has not been well-understood. In this study, the involvement of etiologic factors, such as methylation of FOXP3 gene, was investigated in children with SLE.
MethodTwenty-four female children with SLE and 25 female healthy subjects without any history of autoimmune and inflammatory diseases were included in this study. Blood samples were obtained and DNA was extracted from the blood cells. The bisulphite method was used to convert the DNA using the MethylEdge™ Bisulfite Conversion System Kit. Then, methylation of the gene was investigated using Real Time methylation specific PCR.
ResultsThe FOXP3 DNA methylation in patients and healthy subjects was significantly different. While the median unmethylated DNA in patients was 0.57±0.43, it was 0.97±0.83 in healthy subjects (P=0.012). The Demethylation Index in patients was 0.007±0.003, significantly lower than in controls (0.014±0.013; P=0.012).
ConclusionsThe FOXP3 gene methylation in children with SLE was significantly higher than healthy subjects, which could possibly affect the level of gene expression. Therefore, one of the causes of increased immune response in SLE can be the lower expression of FOXP3 by hypermethylation of this gene.
Considered as a multi-organ auto-immune disease, systemic lupus erythematosus (SLE) affects several body organs such as kidney, joints, lung, nervous system and skin through the production of auto-antibodies against self-antigens.1 Although not clearly understood, loss of tolerance to self-antigens could possibly play a causative role in the pathophysiology of SLE.1 The causative risk factors for SLE could be categorized as genetic and environmental factors in general. While the genes encoding human leukocyte antigen (HLA), cytokines, cell surface antigens and some other genes consistute the genetic arm of SLE risk factors, sex hormones, viral infections, UV radiation, and some medications play causative roles as environmental risk factors.2
Considering the undeniable role of immunity in the pathogenesis of SLE, both B cell and T cell-related immunities actively participate in autoimmunity to self-antigens, leading to systemic organ damage in SLE. As the role of CD4+ T cells in the regulation of B-cell function and production of antibodies, any dysregulation in the process could possibly cause autoimmunity.1 The Forkhead Box P3 (FOXP3) gene is a member of fork-winged helix family of transcription factors with the crucial role in normal maturation and activity of CD4+/CD25+ regulatory T cells (Treg), which are responsible for maintaining tolerance against self-antigens in the body.3 The animal studies have shown different expression levels of FOXP3 gene in different immune cells such as regulatory T cells, CD4+/CD25− T cells, or CD4-/CD8+ T cells, which were closely associated with the function of these cells in production of immune-related molecules, inhibition of autoimmunity and inflammation, or intercellular contacts.4 The three conserved noncoding DNA sequences in FOXP3 gene participate in the FOXP3 expression pattern, which is important in Treg production, division, and differentiation in immune tissues.5 Therefore, the methylation status of FOXP3 gene, especially in the conserved noncoding sequences would influence the phenotype of natural CD4+ Tregs.6
Accordingly, as the undeniable role of Tregs in autoimmunity and the role of FOXP3 gene in phenotype and function of Tregs, the structural and epigenetic changes in this gene could play a role in the pathophysiology of autoimmune diseases such as SLE. One of interesting study involved the evaluation of FOXP3 gene methylation in this disease. To our knowledge, two separate studies, in China and South Africa respectively, demonstrated hypomethylation of this gene in SLE patients.6,7 On the other hand, another investigation among SLE patients indicated that the methylation status of FOXP3 gene was remarkably higher in active cases, whilst inactive cases and healthy subjects had lower levels of DNA methylation in this gene.8
Despite the rich investigations in elucidating the role of autoimmunity in SLE, there is still a lack of evidence regarding the role of the FOXP3 gene structure and function in the pathophysiology of this disease. Therefore, the current case-control study was designed to investigate and compare DNA methylation levels in the FOXP3 promoter between children affected with SLE and healthy controls.
Materials and methodsStudy designBased on the STROBE guidelines for observational studies and in order to investigate the difference between DNA methylation of the FOXP3 promoter in pediatric SLE and healthy controls, this case-control study was designed. The study was conducted in the Research Center for Immunodeficiencies (RCID), Children's Medical Center; and was approved by the Ethics Committee of Tehran University of Medical Sciences (TUMS).
Patient selectionIn the 18-month period of study, pediatric patients who were referred to the Rheumatology Clinic of Children's Medical Center with confirmed diagnosis of pediatric SLE by pediatric rheumatologist were screened for eligibility criteria. Patients under 18 years of age, with no self-history or family history of immunodeficiency or autoimmunity diseases other than SLE were eligible for the study and included. The patients who were either suspicious for SLE or had other concomitant diseases were excluded. The control subjects included healthy subjects with no history of autoimmunity in them or their families, who were not matched by age with the patients. The parents or legal guardians of study subjects were requested to fill in the written informed consent form prior to recruitment and laboratory tests.
Blood sampling and DNA extractionIn brief, 5cc of peripheral blood from all study subjects was stored in −20°C prior to DNA extraction. The DNA from whole blood cells was then extracted using Phenol-Chloroform method as described before.9 The quality of the extracted DNA was evaluated through measurement of optical density and the 260/280 ratio.
DNA treatment and bisulfite conversionPrior to DNA methylation assessment, the extracted genomic DNA was treated though the Bisulfite method using MethylEdge™ Bisulfite Conversion System (Promega, Madison, WI, USA) according to the instructions provided by the manufacturer and was stored at −20°C before PCR. In brief, in the sodium bisulfite treatment method, the unmethylated cytosine residues would be converted to uracil, which would be converted to thiamine following PCR amplification. In this method, the 5-methylcytosine (5mC) of genomic DNA would not be influenced.
DNA methylation assessment using methyl specific PCR (MSP)The real-time quantitative multiplex methylation specific PCR (QM-MSP) method was used to assess the FOXP3 promoter methylation10 in the bisulfite-treated genomic DNA. With considerable sensitivity and specificity, the MethySYBR procedure includes two steps of PCR reactions in general:
- 1)
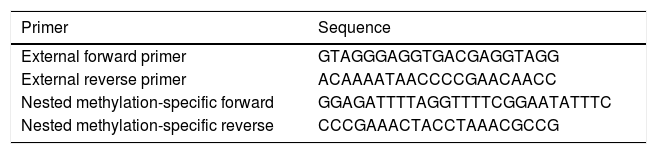
The multiplex step: in this pre-amplification step, paired external primers (external forward primer and external reverse primer) (Table 1) were designed in order to amplify a distinction containing different target alleles. Accordingly, each reaction-well contained a total amount of 25μl mixture including 1μl of converted DNA and other reaction materials. This sequence of reactions included one cycle of 95°C for 5min, and then 30 cycles of 94°C for 30s, 56°C for 30s, and 72°C for 30s. The final cycle of this step was 72°C for 5min.
- 2)
In the second step, the products of the previous step were used to quantify specific methylated targets. In this phase, nested paired primers, independent and specific for methylation, were used. The sequences of nested methylation-specific primers (forward and reverse) are mentioned in Table 1. The UCSC database was used to determine the methylation status of CpG islands in the FOXP3 promoter; and the MethBlast tool was used for primer blasting as well. Each reaction-well contained a total amount of 10μl mixture including 5μl SYBR® Green Master Mix, 0.25μl Forward Primer, 0.25μl Reverse Primer, 3.5μl DDW, and 1μl bisulfite-treated DNA. These reactions started with one cycle of 95°C for one minute, and then 30 cycles of 94°C for 30s, 60°C for 1min, 72°C for 30s, and the final extension of 72°C for 5min.
In order to determine the methylation percentage of DNA samples, a positive MSP control was used in each run which was 100% methylated. For this purpose, fully converted methylated human plasmid DNA was used and therefore, no negative untreated controls were used in this study.
As explained elsewhere, the unmethylated DNA was calculated though the following formulae:11,12
where MSP: methyl specific PCR; BSP: bisulfite specific PCR.Meanwhile, the Demethylation Index (DI) was also calculated as follows, in order to determine the efficiency of the PCR:13
Statistical analysisThe PASS 11 software (version 11.0.4 with serial number 2365456856) was used to calculate the sample size and study power. Group sample sizes of 25 achieve 91% power to detect equivalence when the margin of equivalence is from −1.0 to 1.0 and the actual mean difference is 0.0. The significance level (alpha) is 0.050 using two one-sided Mann–Whitney tests. The amount of unmethylated DNA in each group was reported as median±RIQ; and the difference between groups was assessed using the Mann–Whitney test.
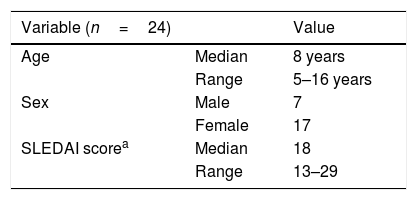
ResultsTwenty-five children with SLE and 25 healthy controls were recruited in this study. After DNA extraction and treatment, the data from 24 patients and 25 controls were analyzable (Table 2).
General description of patients with pediatric systemic lupus erythematosus.
| Variable (n=24) | Value | |
|---|---|---|
| Age | Median | 8 years |
| Range | 5–16 years | |
| Sex | Male | 7 |
| Female | 17 | |
| SLEDAI scorea | Median | 18 |
| Range | 13–29 |
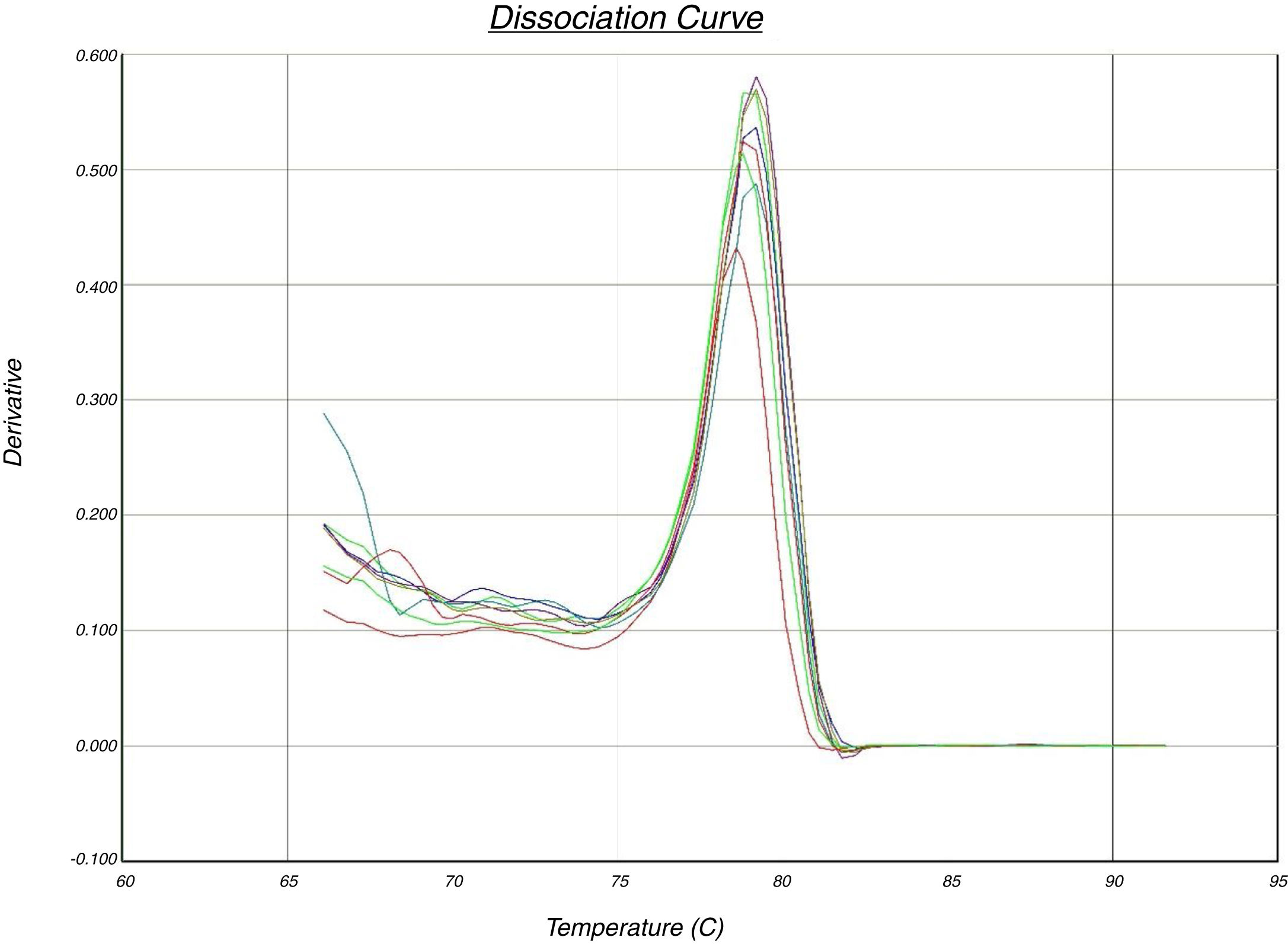
In the quantitative PCR method, the melting curves of target amplicons were analyzed in order to indicate the methylation status of the FOXP3 promoter (Fig. 1).
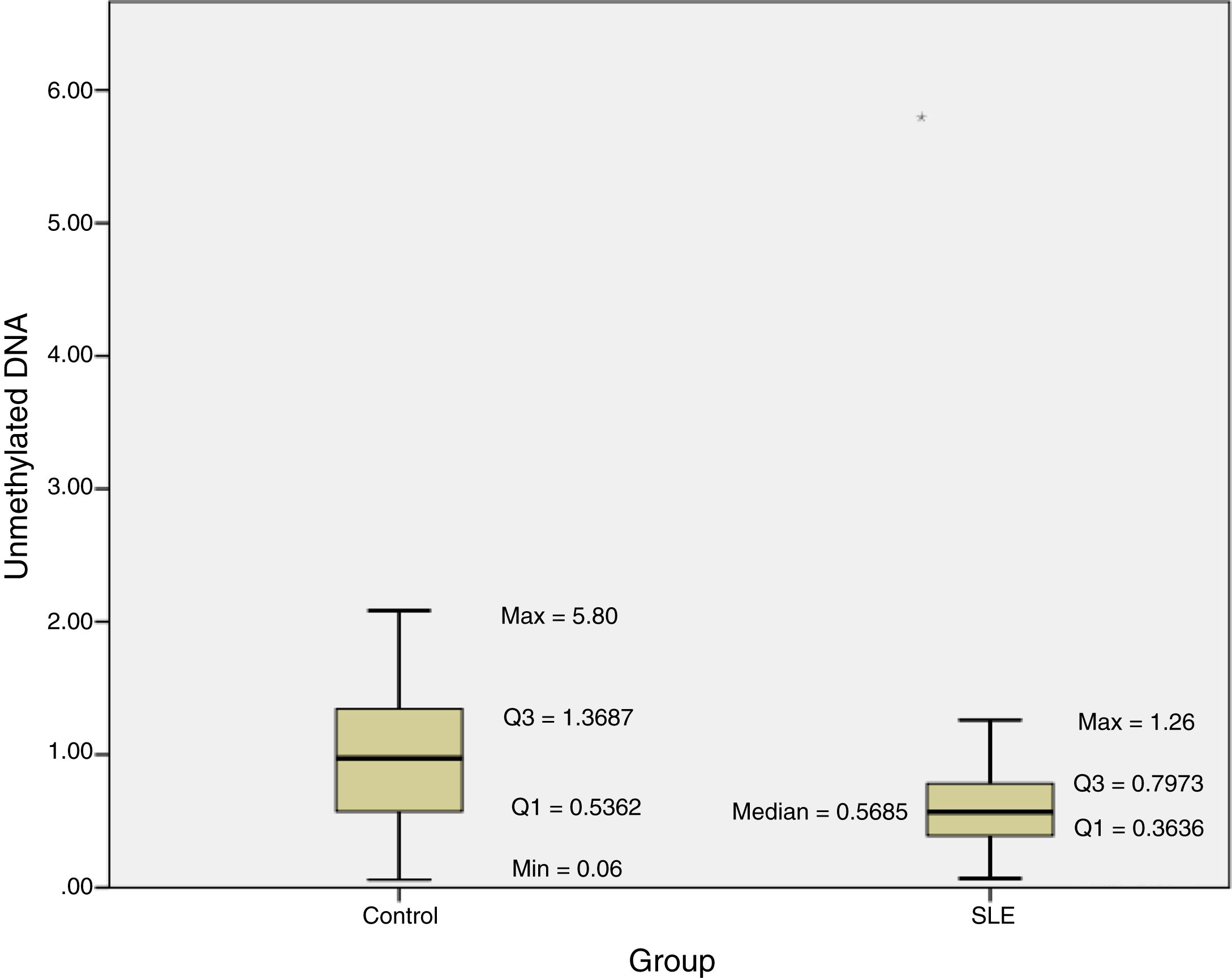
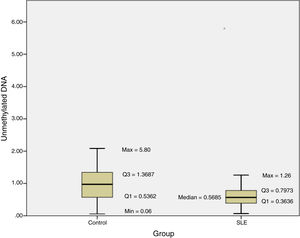
The methylation status of the FOXP3 promoter in patients and controls are shown in Fig. 2. Promoter methylation of the FOXP3 gene was significantly higher in SLE patients; as the median of unmethylated DNA was 0.57±0.43 in the patients, while it was 0.97±0.83 in controls (P=0.012) (Table 3).
Moreover, the DI in the patient group was 0.007±0.003, which was significantly lower than the controls (0.014±0.013; P=0.012) (Table 3).
DiscussionIn order to elucidate the association between promoter methylation of the FOXP3 gene and pediatric SLE, the current investigation was performed to compare the methylation difference between pediatric SLE patients and controls through the Quantitative MSP method.
The results of this investigation indicated significant difference between methylation status of the FOXP3 promoter and SLE as the median of unmethylated DNA was 0.57±0.43 in the patients, while it was 0.97±0.83 in controls (P=0.012).
As a member of the fork-winged helix family (Forkhead), FOXP3 is located at chromosome Xp11.23 and plays an important role in appropriate activity of Tregs, which are important arms in preventing autoimmune reactions. The dysfunction in CD4+/CD25+ T cells, as well as low expression and mutation of the FOXP3 gene were found in association with some autoimmune diseases such as autoimmune hepatitis (AIH), autoimmune thyroid diseases, SLE, rheumatoid arthritis (RA), particular immunodeficiencies, autoimmune diabetes, inflammatory bowel disease (IBD) and many other diseases.14
Several studies investigated the association between this gene and SLE. According to one of these studies, the expression level of two FOXP3 isoforms would be dramatically decreased in patients with SLE and RA.15 On the other hand, another study indicated a significant increase in the FOXP3 presentation level in SLE, with a P-value of 0.012.16 Moreover, the expression of this gene was found associated with renal involvement in SLE and also expression level of anti-dsDNA in these patients as well.17 Playing an important role in the regulation of immune tolerance, the FOXP3+/Helios+ types of regulatory T cells were remarkably more frequent in the blood samples of patients with active SLE (11%) when compared to control subjects (5%) (P<0.001). Importantly, this increase was specifically found in SLE patients and this change was not seen in other autoimmune diseases.18
In addition to an alteration in the expression level of FOXP3 in SLE, different mutations and single nucleotide polymorphisms (SNPs) of this gene were found in association with this disease as well, such as a significant association of rs12843496 of this gene and SLE.19
Similar to the results of association between SLE and expression or mutation of FOXP3, methylation alteration of this gene was also closely associated with SLE and some other autoimmune diseases. Accordingly, in one of these studies, the FOXP3 gene was hypomethylated in SLE patients.6 As investigated among black ethnicity of South Africans, there was a remarkable difference between global methylation in SLE patients and healthy individuals. According to that study, the global methylation was 22.2±5% in patients while it was 33.9±11% in healthy people.7 However, the results of that study were not in concordance with the results of our study, as FOXP3 gene was hypermethylated in the aforementioned patient population.7 However, in another study the mean melting temperature was considered as a criterion for methylation, with a higher mean melting temperature showing higher methylation level. In this study, patients with active SLE disease had a significantly higher methylation level in the FOXP3 gene when compared to both inactive SLE patients and also healthy subjects (79.00±0.28 vs. 78.49±0.29 vs. 78.44±0.25 respectively; P<0.001).8 This result was similar to the findings of the current study.
Moreover, and as primarily described by Akirav et al., the demethylation index mainly compares the relative frequency of unmethylated DNA in cells, which is calculated through subtracting the methylation-specific from bisulfite-specific PCRs.12,13 Accordingly, the difference in the demethylation index between SLE patients and controls was statistically significant, showing lower levels in patients than in controls (P=0.012).
Importantly, the promoter of this gene was demethylated in some other diseases such as IPEX-like syndrome, AIH and RA, resulting in a higher gene expression of FOXP3.20–23 On the other hand, in some other diseases including systemic sclerosis and Sjogren, the promoter of this gene was hypermethylated.24,25
In addition to FOXP3, the methylation alterations of some other genes were also associated with SLE. While demethylation of TNFSF7 was associated with SLE, hypomethylation of some other genes was significantly associated with SLE clinical presentations. Accordingly, hypomethylation of MIR886 and TRIM69 genes were associated with malar rashes, while hypomethylation of PSMB8, RHOJ, and TAP1 were associated with discoid rashes in SLE.26 On the other hand, hypermethylation of some genes such as RUNX3, MHC2TA, RFC1, MBD2, DNMT1, and GR were found correlated to disease as well.27–35
Although the current study provided valuable results in elucidating the pathophysiology of SLE, there were several limitations which shall be considered. First of all was the small sample size, therefore, epidemiologic studies with large sample sizes could be conducted in the future, in order to reduce the possible random error and increase the power. As the clinical and demographic data were not available in this study, several useful analyses measuring the difference between active and inactive phases of disease were not applicable. Therefore, a new prospective study, considering patients’ characteristics is recommended. Meanwhile, the co-effect of other etiologic factors including other genes, age, sex, and environmental factors could be considered as well. As the crucial role of DNA methylation and epigenetics in the expression level and therefore, function of genes, future studies investigating both methylation status and expression level of FOXP3 could more precisely elucidate the role of FOXP3 methylation in the pathophysiology of SLE.
In conclusion, promoter methylation of the FOXP3 gene was significantly higher in pediatric patients with SLE when compared to healthy controls. Therefore, the possible lower expression level of the FOXP3 gene resulted from this hypermethylation could be one of the causes of increased immune response in the pathophysiology of this disease.
Conflict of interestThe authors have no conflict of interest to declare.
The study was supported by a grant from Tehran University of Medical Sciences (95-03-154-32732).