Aspirin-exacerbated respiratory disease (AERD) is a complex clinical syndrome characterised by severe asthmatic attack upon treatment with aspirin and/or non-steroidal anti-inflammatory drugs (NSAIDs). Genetic predisposition has been considered as a crucial determinant and candidate genes have concentrated especially on cysteinyl leukotrienes (LTs)-related genes as the inhibitory action of aspirin and NSAIDs on cyclooxygenase activity may cause overproduction of cysteinyl LTs. However, conflicting results have been reported, in parallel with replication studies in different ethnic groups. Thus, future areas of investigations need to focus on comprehensive approaches towards the discovery of other genetic biomarkers. Unfortunately, few papers have been reported about gene polymorphisms in Japanese patients with AERD. Here, we described on our recent genetic investigations on B2ADR, IL-13, IL-17A, CYP2C19, TBXA2R, CRTH2 and HSP70. This review indicates potential genetic biomarkers contributing to the early diagnosis of AERD, which may include CYP2C19 and HSP70 gene polymorphisms, and future validation studies in independent population are required to provide reassurance about our findings.
Aspirin-exacerbated respiratory disease (AERD), so-called aspirin-intolerant asthma, is an acute asthmatic attack due to ingestion of aspirin and other non-steroidal anti-inflammatory drugs (NSAID). However, the pathophysiological mechanisms underlying the development of this specific asthma phenotype have not yet been fully understood.
Because aspirin intolerance is found only in a specific population, genetic predisposition is considered a crucial determinant for the development of AERD. The inhibitory action of aspirin and NSAID on cyclooxygenase (COX) activity may cause diversion to the 5-lipoxygenase pathway, which leads to the overproduction of cysteinyl leukotrienes (LTs).1 Therefore genetic association studies of LT-related genes have been undertaken to explore the genetic determinants of AERD. In fact, LTC4 synthase promoter polymorphism has been reported to be associated with AERD.2,3 Several investigations have shown that the genetic polymorphisms of 5-lipoxygenase promoter4 and cysteinyl LT receptor 1 promoter5 are risk factors for susceptibility to AERD. However, conflicting results have been reported6,7 indicating that in parallel with replication studies in different ethnic groups, future areas of investigation should focus on the identification of genetic biomarkers for early diagnosis of AERD. In fact, Higashi et al.,8 demonstrated that prostaglandin D2 (PGD2), a major prostanoid synthesised, among other cell types, by activated human mast cells, was overproduced during aspirin-intolerant bronchoconstriction, and no differences in the levels of lipoxygenase products have been found in blood from patients with AERD and those with aspirin-tolerant asthma (ATA).9
In this review, we report on the recent genetic investigations from our laboratory in Japanese patients with AERD,10–14 which was performed with the approval of the Institutional Ethics Committee and with written informed consent from each individual prior to beginning the study. The target DNA sequence of each single-nucleotide polymorphism (SNP) was amplified using a set of primers as shown in each study, and allelic discrimination assay for the target SNP relating to the expression of each gene polymorphism was carried out as shown in the following section. Each study was carried out using the methods described below.
ß2-adrenergic receptor (B2ADR) genes analysisB2ADR is encoded by intronless gene, which is located on chromosome 5q31-32.15 It contains several reported SNPs,16 including Arg16Gly (A46G, rs1042713), Gln27Glu (C79G, rs1042714) and Thr164Ile (C491T, rs1800888).17–19 Although the B2ADR gene is not considered to be a major susceptibility gene for asthma, it has been suggested that its variant alleles may play a role in intermediate or asthma-associated phenotypes,20 such as airway hypersensitivity,21 asthma severity22 and response to specific medications.23
As shown in the previous reports about the genotype frequencies of the B2ADR gene in Asian populations the allelic frequency of Gln27Glu polymorphism of the B2ADR gene is less prevalent among Japanese than in Caucasian population and only 7.5% of the subjects carried the polymorphism.24 In fact, the frequency of Gln27Glu in a Japanese population is 2.3% in the dbSNP database of the National Centre for Biotechnology Information. On the other hand, the frequency of Arg16 allele is 53.8% in a Japanese population, which is similar to that observed in a Caucasian population.24 So, we hypothesised that B2ADR gene polymorphisms might differ between patients with AERD and those with ATA.
DNA in the specimens (from 95 patients with AERD, 300 patients with ATA, and 100 normal controls) obtained by rubbing buccal mucosa with a cotton swab was extracted by using QIAamp 96 DNA blood kits (Qiagen, Hilden, Germany). The target DNA sequence of the B2ADR NM_000024.4 was amplified using a set of primers that were previously described25,26 (forward, nucleotides 188–212: 5′-AGCCAGTGCGCTCAC-CTGCCAGACT-3′; reverse, nucleotides 406–383: 5′-GCTCGAACTTGGCAATGGC-TGTGA-3′) to generate an amplicon of 219bp in length. Allelic discrimination assay for SNPs relating to the B2ADR expression (rs 1042713) was carried out using previously described SNPs detective system, sequence-specific thermal-elution chromatography.27 All subjects and investigators remained unaware of the genotype until the final analysis. Allele frequencies were estimated by gene counting method. Significant departures of genotype frequency from the Hardy–Weinberg equilibrium were tested by the Chi-square analysis. Differences in minor allele (Gly) frequency in patients with AERD and control subjects were compared with that in patients with ATA by means of the Chi-square test and calculation of odds ratio (OR) with 95% confidence interval (CI). OR with 95% CI associated with ArgArg of patients with AERD was compared with that of patients with ATA. Polymorphisms related to the asthma phenotype were further examined by multivariable logistic regression analysis with adjustment for covariates. Statistical analyses were undertaken using SPSS for Windows version 17 (SPSS Inc, Chicago, IL, USA).
We10 showed that the frequencies of wild-type ArgArg homozygote were significantly higher than those of variant-type ArgGly/GlyGly genotype in patients with AERD compared to those with ATA (p<0.001), and the OR of patients with AERD associated with wild-type ArgArg homozygote to those with variant-type ArgGly/GlyGly genotype was 3.153 (95% CI=1.789–5.558). In patients with AERD, frequencies of wild-type ArgArg homozygote in both female and male patients were significantly higher than those of variant-type ArgGly/GlyGly genotype in male patients compared with those with ATA (p<0.001, OR=5.128, 95% CI=2.331–11.236 in female and p=0.007, OR=4.367, 95% CI=1.495–12.821 in male, respectively). Also, in patients with AERD, frequencies of wild-type ArgArg homozygote in female patients were significantly higher than those of variant-type ArgGly/GlyGly genotype in female patients compared to those with ATA (p=0.002, OR=2.825, 95% CI=1.453–5.495).
A study from Korea indicated a possible interaction of four loci including Arg16Gly genotype and cysteinyl LT receptor 1 promoter genotype in Korean subjects with AERD,28 suggesting the possible interactions with B2ADR and overproduction of cysteinyl LTs in pathobiology of AERD. So, further studies are needed in Japanese people.
Cytokine genes analysisInterleukin-13 (IL-13), mainly but not exclusively produced by TH2 lymphocytes, is well known to be involved in eosinophilia and airway hyperresponsiveness.29 On the other hand, it has been demonstrated that human TH17 cells, like in mice,30 express IL-13 ¿1-receptor and that IL-13 attenuates IL-17A production.31
The IL-13 gene is located on chromosome 5q31-33, a region frequently linked to asthma.32,33 Two of the most characterised SNPs in IL-13 include a promoter SNP (−1111C>T) and a coding SNP in exon 4 (Arg130Gln). The IL-13 Arg130Gln polymorphism is associated with an elevated eosinophil count and high total serum IgE levels.34–36 Functional studies support a regulatory role associated with allergic inflammation for the −1111C>T variant.36,37
The IL-17A gene is located on chromosome 6q12.1, a genomic region associated with different types of asthma.38–40 A study on the association between asthma susceptibility and IL-17A gene polymorphisms in a Taiwanese population has shown that among nine SNPs investigated, only one SNP (−737C>T) was associated with asthma, and the risk genotype of the SNP was the CC genotype.41 So, we hypothesised that IL-13 and IL-17A gene polymorphisms might be involved in susceptibility to AERD, and performed the study using the DNA specimens obtained from 95 patients with AERD, 300 patients with ATA, and 100 normal controls. The target DNA sequence of the IL-13 −1111C>T was amplified using a set of primers (forward: 5′-TGGGGGTTTCTGGAGGAC-3′, reverse: 5′-GCAGAATGAGTGCTGTGGAG-3′) and that of Arg110Gln was amplified using a set of primers (forward: 5′-GGTCCTGTCTCTGCAAATAATG-3′, reverse: 5′-GTTTTCCAGCTTGCATGTCC-3′). The target DNA sequence of the IL-17A −737C>T was amplified using a set of primers (forward: 5′-CCCCCATCATGT-CTCCTCTCC-3′, reverse: 5′-CCAAGCAACTTGGTGTTTTGAGG-3′). Allelic discrimination assay for SNPs relating to the expressions of IL-13 −1111C>T, IL-13 Arg110Gln and IL-17A −737C>T (rs1800925, rs20541 and rs8193036, respectively) was carried out.
We11 showed that the frequencies of the combined homozygous TT and heterozygous CT genotype group of IL-13 −1111C>T were higher than those of the homozygous CC genotype in AERD patients compared to those with ATA (p<0.001), and the OR of patients with AERD associated with the combined TT/CT genotype group to those with CC genotype was 2.818 (95% CI=1.727–4.597). A positive association between asthma phenotype and the IL-13 −1111C>T genotype was found in female patients, and the frequencies of the combined TT/CT genotype group in female patients with AERD were higher than those of the CC genotype group compared to female ATA patients (p<0.001, OR=3.505, 95% CI=1.946–6.312). No association between asthma phenotype and the IL-13 Arg130Gln genotype was found. The frequencies of the CC genotype of IL-17A −737C>T were higher than those of the combined TT/CT genotype group in AERD patients compared to ATA patients (p=0.015, OR=1.797, 95% CI=1.123–2.877). A positive association between asthma phenotype and the genotype was present in female patients, and the frequencies of the CC genotype in female patients with AERD were higher than those of the combined TT/CT genotype group compared to those with ATA (p=0.030, OR=1.857, 95% CI=1.063–3.244). Comparison of the clinical characteristics in AERD patients according to the IL-13 and IL-17A gene polymorphisms revealed that FEV1 in the patients with the homozygous CC genotype of the IL-13 −1111C>T gene was lower than that in the patients with the combined TT/CT genotype group (p=0.048). AERD patients with the CC genotype of the IL-17A −737C>T gene had a lower peripheral total eosinophil count than did the patients in the combined TT/CT genotype group (p=0.033).
As far as the authors investigated, only one study has reported the association between the IL-13 gene polymorphism and AERD, the findings of which indicated that the allele and genotype frequencies of two promoter polymorphisms of the IL-13 −1510A>C and −1055C>T gene and Arg110Gln polymorphism were not associated with AERD in a Korean population.42 However, to our knowledge, no studies have evaluated IL-17A gene polymorphism association with AERD. So, with the results, we hypothesise that the interaction between IL-13 −1111C>T and IL-17A −737C>T gene sequence variations might be involved in the process to induce allergic inflammation associated with eosinophilic inflammation in AERD.
Cytochrome P450 genes analysisPolymorphisms of the cytochrome P450 (CYP) gene, including CYP2C9 and CYP2C19, have major consequences on the metabolism of a variety of drugs. NSAIDs are metabolised by CYP2C9 in vitro, and the CYP2C9 genotype was considered to be a relevant risk factor for side effects. However, the CYP2C9 genotype has no clinically meaningful effect on the pharmacokinetics of NSAIDs.43–45
The CYP2C19 gene is located on chromosome 10, and two major SNPs are known to make the enzyme activity non-functional.46,47 One is CYP2C19*2 at position 681 in exon 5 (681 G>A), and the other is CYP2C19*3 at position 636 in exon 4 (636 G>A). The polymorphism of this enzyme leads to patient classification into three distinct groups: rapid metaboliser (RM: *1/*1), intermediate metaboliser (IM: *1/*X) and poor metaboliser (PM: *X/*X; *1 and *X represent the wild-type and mutant allele, respectively).
The association between the CYP2C19*2 polymorphism and inflammatory maker concentrations has been reported, and the polymorphism of the CYP2C19 gene might be considered a new candidate for cardiovascular risks through inflammation.48 CYP2C19 has endogenous substrates, including arachidonic acid (AA), such as hydroxyeicosatetraenoic acids (HETEs).49 Therefore we hypothesised that the CYP2C19 gene polymorphism might be involved in the susceptibility to AERD, and performed the study using the DNA specimens obtained from 100 patients with AERD, 300 patients with ATA, and 100 normal controls. The target DNA sequence of CYP2C19 681G>A was amplified using a set of primers (forward: 5′-TTTCCCACTATCATTGATTATTTCC-3′, reverse: 5′-TCTCCATTTTGATCAGGAAGC-3′). The target DNA sequence of CYP2C19 636G>A was amplified using a set of primers (forward: 5′-TGAAAACATCAGGATTGTAAGCAC-3′, reverse: 5′-ATATTCACCCCATGGC-TGTC-3′). Allelic discrimination assay for SNPs relating to the expressions of CYP2C19 681G>A and 636G>A (rs4244285 and rs4986893, respectively) was carried out.
We12 showed that the frequencies of two alleles, *2 and *3, were higher than those of the *1 allele in patients with AERD compared to those seen in patients with ATA and healthy control subjects (p<0.001). The frequencies of PM (*2/*2, *2/*3, *3/*3) were higher than those of RM (*1/*1) and IM (*1/*2, *1/*3) in patients with AERD compared to those seen in patients with ATA (p<0.001). The frequencies of IM and PM were higher than those of RM in patients with AERD compared to those seen in patients with ATA (p=0.001). The frequencies of PM were higher than those of RM and IM in patients with AERD compared to those seen in healthy control subjects (p<0.001). The frequencies of IM and PM were higher than those of RM in patients with AERD compared to those seen in healthy control subjects (p<0.001).
The frequencies of the combined GA/AA genotype group of CYP2C19 681G>A gene were higher than those of GG in patients with AERD compared to those seen in patients with ATA (p=0.001, OR=2.250, 95% CI=1.407–3.598), and the frequencies of the combined GA/AA genotype group of CYP2C19 636G>A gene were higher than those of GG in patients with AERD compared to those seen in patients with ATA (p<0.001, OR=3.694, 95% CI=2.297–5.940). The frequencies of the combined GA/AA genotype group of CYP2C19 681G>A gene were higher than those of GG in patients with AERD compared to those seen in patients with ATA (p=0.008, OR=3.621, 95% CI=1.402–9.351 in male, and p=0.022, OR=1.899, 95% CI=1.099–3.281 in female, respectively). The frequencies of the combined GA/AA genotype group of CYP2C19 636G>A gene were higher than those of GG in patients with AERD compared to those in patients with ATA (p<0.001, OR=7.671, 95% CI=2.913–20.202 in male, and p<0.001, OR=2.822, 95% CI=1.624–4.903 in female, respectively). Finally, the comparison of the clinical characteristics according to CYP2C19 681G>A and 636G>A gene polymorphisms in patients with AERD showed that percent predicted FEV1 in AERD patients with the GG genotype of each CYP2C19 gene were higher than that seen in patients with the combined GA/AA genotype group (p<0.001).
CYP2C19 has been known to be highly implicated in the metabolic turnover of AA, and the functional enzyme product of the CYP2C19*1 oxygenates AA to various HETE metabolites, even though the relevance of the CYP2C19 polymorphism in the production of AA metabolites in the inflammation-linked diseases has been poorly documented. Recently, the association between the CYP2C19*2 allele and inflammatory maker concentrations has been reported.48 We first investigated the frequencies of the CYP2C19 681G>A and 636G>A genotype in AERD patients with our hypothesis that this mutant allele could also be involved in a defect in AA metabolism, leading to its accumulation and thus indirectly to the inflammatory reaction in patients with AERD. Indeed, a specific aspirin-triggered enhancement of 15-HETE generation from nasal polyp epithelial cells and peripheral blood leukocytes from patients with AERD, but not from patients with ATA, has been demonstrated.50–52 On the other hand, the CYP2C19*2 allele has been shown to be associated with higher platelet aggregability,53 which might modify thromboxane (TX) production from platelets. Also, it has been demonstrated that aspirin led to a significant decrease in serum TXB2 levels in patients with persistent asthma.54 Taking these reports into consideration, our data suggest that CYP2C19 gene polymorphism profiles may be a useful diagnostic tool in assessment of the susceptibility to AERD.
Prostanoids-related genes analysisThe TBXA2 receptor (TBXA2R) gene exists on chromosome 19p13.3, and conflicting results about the genetic alteration of TBXA2R in the involvement of asthma have been reported in a Korean population. Namely, Shin et al.,55 showed a positive association between the TBXA2R polymorphism and the development of atopy and asthma. On the other hand, Kim et al.,56 showed that the TBXA2R polymorphism was not associated with asthma susceptibility and the clinical parameters of asthma. In a Japanese population, Unoki et al.,57 found that the synonymous +924T>C polymorphism in the TBXA2R gene was associated with a diagnosis of asthma in adult asthmatic patients, but not in children. However, their claim of an involvement of TBXA2R in Japanese asthmatics does not seem to be substantiated by their data.58,59
The gene of the human chemoattractant receptor expressed on type 2 helper T cells (CRTH2), a receptor for PGD2, is located on chromosome 11q13 and genetic alteration of CRTH2 has been associated with allergic asthma in African-American and Chinese populations.60 However, no association has been found between any polymorphisms or haplotypes in the CRTH2 gene and asthma in the Japanese population.61 So, we hypothesised that TBXA2R and CRTH2 gene polymorphisms might be involved in susceptibility to AERD, and performed the study using the DNA specimens obtained from 96 patients with AERD, 500 patients with ATA, and 100 normal controls. The target DNA sequence of TBXA2R +795T>C was amplified using a set of primers (forward: 5′-GAGTGGACCCTGGATCTCAA-3′, reverse: 5′-CCACGCGCAAGTAGATGAG-3′). The target DNA sequence of CRTH2 −466T>C was amplified using a set of primers (forward: 5′-GAGCTGCATGGAGGATCTGT-3′, reverse: 5′-AGGACTCC-TTTTTCCCATCC-3′). Allelic discrimination assay for SNPs relating to the expressions of TBXA2R +795T>C and CRTH2 −466T>C (rs11085026 and rs634681, respectively) was carried out.
We13 showed that the frequencies of the combined CC/CT genotype group of the TBXA2R +795T>C were significantly higher than those of the homozygous TT genotype in patients with AERD compared to those in patients with ATA (p=0.015, OR=1.748, 95% CI=1.116–2.739). The frequencies of the combined CC/CT genotype group of the TBXA2R +795T>C were significantly higher than those of the homozygous TT genotype in female patients with AERD compared to those in female patients with ATA (p=0.013, OR=1.961, 95% CI=1.150–3.346). The frequencies of the homozygous TT genotype of the CRTH2 −466T>C in patients with AERD were significantly higher than those of the combined CC/CT genotype group compared to those in patients with ATA (p=0.034, OR=1.616, 95% CI=1.037–2.518). The frequencies of the homozygous TT genotype of the CRTH2 −466T>C in female patients with AERD were significantly higher than those of the combined CC/CT genotype group compared to those in female patients with ATA (p=0.046, OR=1.712, 95% CI=1.010–2.903).
Investigations of the association between AERD susceptibility and prostanoid gene polymorphisms in a Korean population have shown that, among three SNPs of the TBXA2R gene investigated, the +795T>C polymorphism was only associated with AERD susceptibility62,63 and the −466T>C polymorphism of the CRTH2 gene was associated with AERD.64 However, there has been no published data addressing the role of TBXA2R and CRTH2 gene polymorphisms in Japanese patients with AERD.
In our study, the relationship between the genotyping and clinical findings in patients with AERD was not demonstrated. Notably, an agonistic effect of indomethacin on a CRTH2 has been reported,65 which may lead to eosinophilic infiltration in AERD patients. Nevertheless, it is easy to speculate that a single genetic factor cannot explain the genetic background of AERD.
Heat shock protein (HSP) genes analysisHSP, of which the HSP70 family is best understood, responds to a variety of stressful stimuli by augmentation of its intracellular HSP gene expression66 and subsequent inhibition of pro-inflammatory cellular functions.67
There are three genes in the HSP70 family HSPA1A, HSPA1B and HSPA1L, located adjacent to each other in the class III region of the major histocompatibility complex (MHC) (chromosome 6p21.3).68 The two intronless genes, HSPA1A and HSPA1B, encode an identical protein.69 Both genes are expressed at high level in cells upon heat shock, with HSPA1A also expressed constitutively at very low levels.68 The HSPA1L gene is expressed at low levels both constitutively and following heat shock.70
A prospective cohort study of community-acquired pneumonia found that carriage of the AA homozygotes of HSPA1B1267A>G gene was associated to a significantly greater risk of developing septic shock.71 As HSPA1B1267A>G is a silent mutation, it is likely that another polymorphic site is responsible for the changes in biological function that explain the disease association. In fact, HSPA1B1267A>G and HSPA1B-179C>T was found to be in linkage disequilibrium.71 Temple et al.,72 investigated the promoter region of HSPA1A and HSPA1B in healthy whites and Asians, and demonstrated that HSPA1B-179C>T is in linkage disequilibrium with HSPA1B1267A>G, and HSPA1B-179C>T affects HSP70 production, suggesting HSPA1B-179C>T as a key determinant of individual susceptibility to a variety of inflammatory diseases. The data were sub-analysed by race, and the same associations were observed in Caucasians and Asians. They also suggested HSPA1B-179C>T:1276A>G haplotype is functional and may explain the association of the HSP70 gene with development of septic shock.73 So, we hypothesised that HSPA1B-179C>T and 1267A>G gene polymorphisms might be involved in the susceptibility to AERD, and performed the study using the DNA specimens obtained from 102 patients with AERD, 300 patients with ATA, and 100 normal controls. The target DNA sequence of the HSPA1B-179C>T was amplified using a set of primers (forward: 5′-AAAGGCGGGT-CTCCACGAC-3′, reverse: 5′-GTTCGCGCTCTGGAAAGCCTTG-3′) and that of 1267A>G was amplified using a set of primers (forward: 5′-ACAAGGCCCAGATTC-ACG-3′, reverse: 5′-GTTTTCCAGCTTGCATGTCC-3′). Allelic discrimination assay for SNPs relating to the expressions of HSPA1B-179C>T and 1267A>G (rs6457452 and rs1061581, respectively) was carried out. Differences in the clinical characteristics according to the association of the HSPA1B-179C>T and 1267A>G gene polymorphisms in the patients were compared by the F-test, and qualitative data were compared by the Chi-square test. Linkage disequilibrium (LD) evaluated by D′ coefficient was calculated to evaluate linkage disequilibrium by Haploview version 4.0 software programme. Haplotype frequency analysis with multivariate adjustment for age and gender was determined using the H-Plus haplotype version 2.5 software programme.74,75
We14 showed that AERD patients showed higher frequencies of combined CT/TT genotype group of the HSPA1B-179C>T than those of homozygous CC genotype compared to ATA patients (p<0.001, OR=7.527, 95% CI=3.933–14.407), and higher frequencies of homozygous GG genotype of the HSPA1B1267A>G than those of combined GA/AA genotype group compared to ATA patients (p<0.001, OR=3.126, 95% CI=1.953–5.001). Also, AERD patients showed higher frequencies of combined CT/TT genotype group of the HSPA1B-179C>T than those of CC genotype compared to ATA patients (p=0.001, OR=8.500, 95% CI=2.297–31.45 in male and p<0.001, OR=7.382, 95% CI=3.459–15.754 in female, respectively), and higher frequencies of homozygous GG genotype of the HSPA1B1267A>G than those of combined GA/AA genotype group compared to ATA patients (p=0.022, OR=2.758, 95% CI=1.161–6.550 in male and p<0.001, OR=3.308, 95% CI=1.887–5.769 in female, respectively). On the basis of the LD coefficient (D′=1.000) between the two genotype SNPs in normal controls in the present study, we inferred the haplotype frequencies and showed that the prevalence of haplotype [C-A] was significantly higher in AERD patients than in ATA patients (p<0.001, OR=3.154, 95% CI=1.916–5.193). Investigations about the relationship between the haematological characteristics and the association of HSPA1B-179C>T and HSP1267A>G gene polymorphisms showed a significant variance in peripheral blood total eosinophil count in AERD patients (p=0.033), but not in ATA patients.
Aron et al.,76 suggested that HSP70 overexpression in asthma was independent on HSP gene polymorphisms, and Smith et al.,77 reported HSPA1A and HSPA1B do not share common patterns of polymorphisms. HSPA1B1267A>G is a silent polymorphism,78 and it is likely that another polymorphic site is involved in the biological function and explains the disease association. In fact, it has been reported that HSPA1B-179C>T is in linkage disequilibrium with HSPA1B1267A>G, and the A allele of HSPA1B1267 is in linkage with the C allele of the HSPA1B179 which is associated with lower levels of gene expression,71 suggesting that HSPA1B gene polymorphism is one of the key determinants of individual susceptibility to a variety of infectious and inflammatory diseases.
AERD is known to be associated with higher peripheral blood eosinophil count than ATA,79,80 which corresponds to the results in our investigations.11–14 We first investigated the frequencies of the HSPA1B-179C>T and 1267A>G genotype in AERD patients and ATA patients, and demonstrated that the prevalence of haplotype [C-A] was significantly higher in AERD patients than in ATA patients. While studies investigating the levels of translated protein still need to be performed, the A allele of HSPA1B1267 has been shown to be in linkage with the C allele of the HSPA1B179 which is associated with lower levels of HSP70 gene expression.71 So, a lower production of intracellular HSP70 may have a minimal effect on inhibiting pro-inflammatory cellular functions potentially involved in AERD. The present study also provides evidence, a significantly elevated peripheral blood total eosinophil count associated to the two SNPs in AERD patients, but not in ATA patients, indicating that the association with HSPA1B-179C>T and 1267A>G gene polymorphisms may be involved in the susceptibility to AERD.
Studies in cultured cells have demonstrated NSAIDs can potentiate heat-induced HSP70 expression,81 however the use of NSAIDs has been recommended to be carefully monitored in cancer patients undergoing hyperthermic treatment.82,83 On the other hand, Mortaz et al.,84 demonstrated that NSAIDs induced HSP70 from bone marrow-derived mast cells was closely paralleled with inhibition of tumour necrosis factor (TNF) production. They also demonstrated that aspirin-induced release of HSP70 from mast cells results in cell activation through Toll-like receptor pathway.85 Interestingly, Kee et al.,73 found that individuals with the haplotype containing the sepsis-associated genotype, HSPA1B-179*C:HSPA1B1267*A, have decreased expression of HSP70 in mononuclear cells and increased production of TNF. TNF is a well-known pro-inflammatory cytokine released from inflammatory cells including mast cells,86,87 and is increased in asthmatic airways.88 As the HSP genes lie in the MHC class III region,68 it is possible that linkage of HSPA1B-179C>T with other polymorphisms in this region and the adjacent TNF genes may account for some of the functional associations. Because many articles have indicated that mast cells may be involved in the pathogenesis of AERD89–91 including that by Higashi et al.,8 the results of the present study may suggest a role of mast cells in AERD through aspects of HSP70 as proposed.92 In fact, Zander et al.,93 demonstrated that protein microassay analysis of nasal polyps from aspirin-sensitive patients with chronic rhinosinusitis showed a greater expression of HSP70 than that from aspirin-tolerant patients. Taking these reports into consideration, our data suggest that HSP70 gene polymorphism profiles may be a useful diagnostic tool in assessment of the susceptibility to AERD.
ConclusionsAERD often produces moderate-to-severe phenotype asthma, however diagnosis of AERD is challenging despite the availability of various techniques including lysine-aspirin challenge test. A hypothesis has been put forward, mostly focused on the overproduction of cysteinyl LTs. Because aspirin intolerance is found in a specific population, genetic predisposition is considered a crucial determinant for its development. Although candidate studies have concentrated on the cysteinyl LT-related genes, conflicting results have been reported, and further areas of genomic investigations need to be focused on identification of biomarkers for the early diagnosis of AERD. In this review, we described the recent genetic investigations from our laboratory. We were the first to analyse the CYP2C19 and HSP70 gene polymorphisms in AERD patients. Our data suggest that CYP2C19 and HSP70 gene sequence variations might be implicated in the development of AERD, and these gene polymorphism profiles may be a useful diagnostic tool in assessment of the susceptibility to AERD patients (Table 1). The findings of our studies were based on small-sized samples from a Japanese population, and further validation studies in independent populations are thus required to confirm the findings.
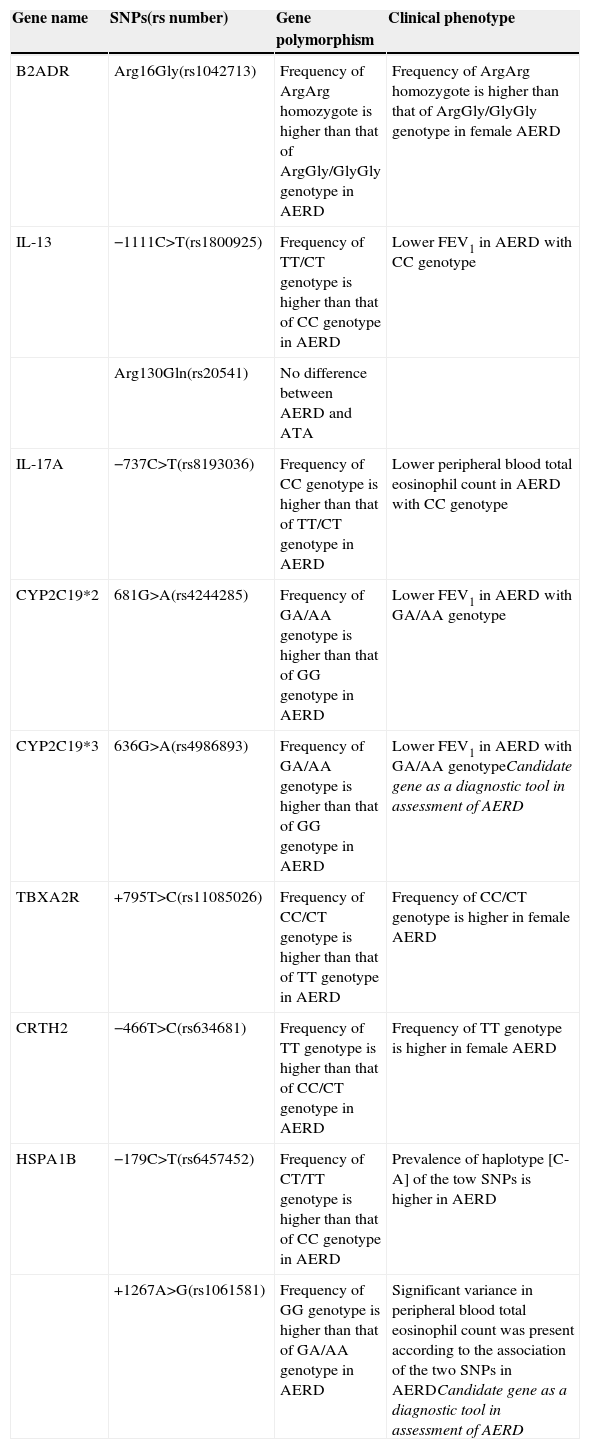
Candidates of gene polymorphisms involved in clinical phenotype of AERD.
| Gene name | SNPs(rs number) | Gene polymorphism | Clinical phenotype |
|---|---|---|---|
| B2ADR | Arg16Gly(rs1042713) | Frequency of ArgArg homozygote is higher than that of ArgGly/GlyGly genotype in AERD | Frequency of ArgArg homozygote is higher than that of ArgGly/GlyGly genotype in female AERD |
| IL-13 | −1111C>T(rs1800925) | Frequency of TT/CT genotype is higher than that of CC genotype in AERD | Lower FEV1 in AERD with CC genotype |
| Arg130Gln(rs20541) | No difference between AERD and ATA | ||
| IL-17A | −737C>T(rs8193036) | Frequency of CC genotype is higher than that of TT/CT genotype in AERD | Lower peripheral blood total eosinophil count in AERD with CC genotype |
| CYP2C19*2 | 681G>A(rs4244285) | Frequency of GA/AA genotype is higher than that of GG genotype in AERD | Lower FEV1 in AERD with GA/AA genotype |
| CYP2C19*3 | 636G>A(rs4986893) | Frequency of GA/AA genotype is higher than that of GG genotype in AERD | Lower FEV1 in AERD with GA/AA genotypeCandidate gene as a diagnostic tool in assessment of AERD |
| TBXA2R | +795T>C(rs11085026) | Frequency of CC/CT genotype is higher than that of TT genotype in AERD | Frequency of CC/CT genotype is higher in female AERD |
| CRTH2 | −466T>C(rs634681) | Frequency of TT genotype is higher than that of CC/CT genotype in AERD | Frequency of TT genotype is higher in female AERD |
| HSPA1B | −179C>T(rs6457452) | Frequency of CT/TT genotype is higher than that of CC genotype in AERD | Prevalence of haplotype [C-A] of the tow SNPs is higher in AERD |
| +1267A>G(rs1061581) | Frequency of GG genotype is higher than that of GA/AA genotype in AERD | Significant variance in peripheral blood total eosinophil count was present according to the association of the two SNPs in AERDCandidate gene as a diagnostic tool in assessment of AERD |
Confidentiality of data. The authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflicts of interestAuthors have no competitive or financial interests to disclose. No support was provided for the present study.