Pinus subsection Australes is a group of North American hard pines comprising approximately 29 ecologically and economically important tree species distributed throughout North and Central America and the Caribbean Islands. Previous studies have shown that some species of this subsection share plastid DNA haplotypes, a pattern that is attributed to introgressive hybridization or the retention of ancestral polymorphisms. Here we describe the morphological and plastid haplotype diversity for this group of species in the states of Guerrero and Oaxaca, Mexico. Seven species of Pinus subsection Australes are recognized in the study area, one of which, P. patula, includes two varieties. Seven variable sites and nine haplotypes were found in an 840 b.p. fragment of the DNA coding region ycf1. Shared haplotypes were found for P. patula var. patula, P. patula var. longipedunculata, P. herrerae, and P. tecunumanii. Four of the nine haplotypes found were restricted to Oaxaca. Although plastid DNA genealogies are valuable for studying evolution in this group, greater sampling of individuals and the inclusion of more variable sites are needed to more accurately infer species relationships.
Pinus subsección Australes es un grupo de pinos duros de América del Norte que comprende aproximadamente 29 especies de árboles importantes económicamente y ecológicamente distribuidos a lo largo de toda América del Norte y Central y las Islas Caribeñas. Estudios previos han mostrado que las especies de esta subsección a menudo comparten haplotipos de ADN de plastidio, un patrón que es atribuido a la hibridación introgresiva y la retención de polimorfismos ancestrales. Aquí describimos la diversidad de haplotipos de plastidio y la morfología para este grupo de especies en los estados de Guerrero y Oaxaca, México. Siete especies de Pinus subsección Australes son reconocidas en el área de estudio, una de las cuales, P. patula incluye dos variedades. Siete sitios variables y nueve haplotipos fueron encontrados amplificando un fragmento de 840 p. b. de ADN de la región codificante ycf1. Se encontraron haplotipos compartidos para P. patula var. patula, P. patula var. longipedunculata, P. herrerae y P. tecunumanii. Cuatro de los nueve haplotipos encontrados están restringidos a Oaxaca. Aunque las genealogías de genes son valiosas para estudiar la evolución de este grupo, se requieren mayor muestreo de individuos y más sitios variables para la inferencia de relaciones entre las especies.
Pinus (Pinaceae), arguably the most ecologically and economically important tree genus in the world1, comprises approximately 110 species distributed naturally in terrestrial environments throughout the Northern Hemisphere2–5. Pines are easily recognized by their needle-like leaves arranged in fascicles of 2–8 (although P. monophylla Torr. & Frém. and P. californiarum D.K. Bailey have solitary needles), and woody seed cones with thickened scale apices and a dorsal or terminal umbo (a specialized raised region resulting from more than one season of growth). Mexico is the center of species diversity for pines, with more than 40% of the world's species occurring naturally within its borders2–7. The number of species recognized for the country has varied among recent authors; Eckenwalder2 recognized 39 species, Farjon3 recognized 44, and Gernandt and Pérez-de la Rosa7 recognized 49.
Pinus is classified in two subgenera, four sections, and 11 subsections8. Pinus subsection Australes is the most species rich, comprising approximately 29 species distributed throughout North and Central America and the westernmost islands of the Caribbean except Jamaica. It includes the southern yellow pines, the egg-cone pines, and the California closed-cone pines, and is classified together with subsections Contortae, and Ponderosae in Pinus subgenus Pinus section Trifoliae, a group known informally as the North American hard pines9. All species of Pinus subsection Australes have two vascular bundles in their leaves, and most have persistent fascicle sheaths. All have woody seed cones, and some have strongly developed cone scale apophyses and umbos, often with a mucro. Their seeds have an articulate wing and are primarily wind-dispersed.
Numerous taxonomic works have treated Pinus2,9–11, including regional floras that include Mexico6,12–14. Morphology alone has not been sufficient to resolve the taxonomic questions surrounding the genus. DNA sequences have been very effective at confirming or rejecting the major pine lineages that were originally proposed based primarily on morphology, artificial crosses, and biochemical studies9,15. However, natural and artificial hybridization are relatively well documented in pines16. Both hybridization and retention of ancestral alleles have obscured the phylogenetic relationships among closely related species17–24.
In most organisms, speciation both generates biological diversity and gives rise to gene genealogies that reflect the history of divergence of the organisms in which the genes are found. These histories can be reconstructed using phylogenetic analysis and used to infer the phylogenetic relationships among species. In pines, hybridization and incomplete lineage sorting cause a remarkable disconnection between individual gene trees and species relationships24. As a result, many gene trees need to be considered separately (although not in isolation) to form hypotheses regarding species relationships.
Plastid (e.g., chloroplast) DNA has been widely studied in pines. In contrast to angiosperms, plastid DNA is paternally inherited in conifers25. It exhibits low genetic differentiation in conifer populations and species relative to mitochondrial or nuclear DNA; this is because plastid gene flow is mediated by both seed and pollen in conifers26. One result is that species take a long time to become fixed for a single plastid DNA lineage, and sharing of plastid lineages by closely related species is often observed18,27.
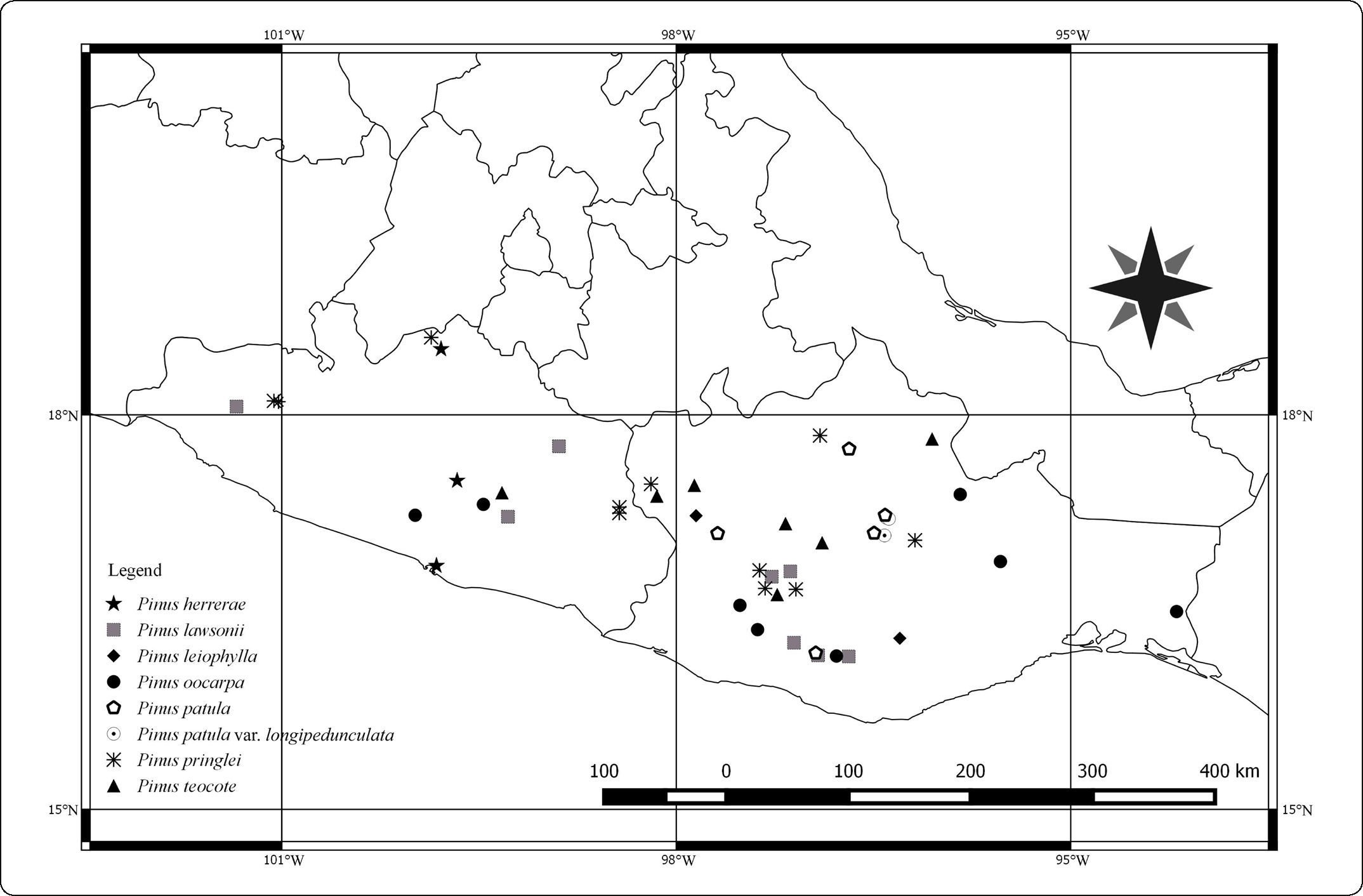
We chose as our study area the political boundaries marked by Guerrero and Oaxaca. These states are located on the southern Pacific coast of Mexico. Guerrero occupies 63,596km2 and is the fourteenth largest state in the country, and Oaxaca occupies 93,757km2 and is the fifth largest28. The principal mountain range in these states is the Sierra Madre del Sur, which extends through Jalisco, Colima, southern Michoacán, the State of Mexico, Morelos, Puebla, Guerrero, and Oaxaca, and is bordered to the north by the Transverse Mexican Volcanic Belt29. Both Guerrero and Oaxaca are highly diverse in species. For example, 9,362 species of plants have been reported for Oaxaca, more than any other state in the country30. Fourteen pine species have been recognized for Guerrero31 and 17 for Oaxaca32. Seven of these species belong to Pinus subsection Australes.
This is the first comparative study to integrate morphological, anatomical, and molecular characters in pines of Guerrero and Oaxaca, Mexico. Our objectives are to better understand the morphological diversity of Pinus subsection Australes in these two states and to further document which species in this group share plastid DNA haplotypes.
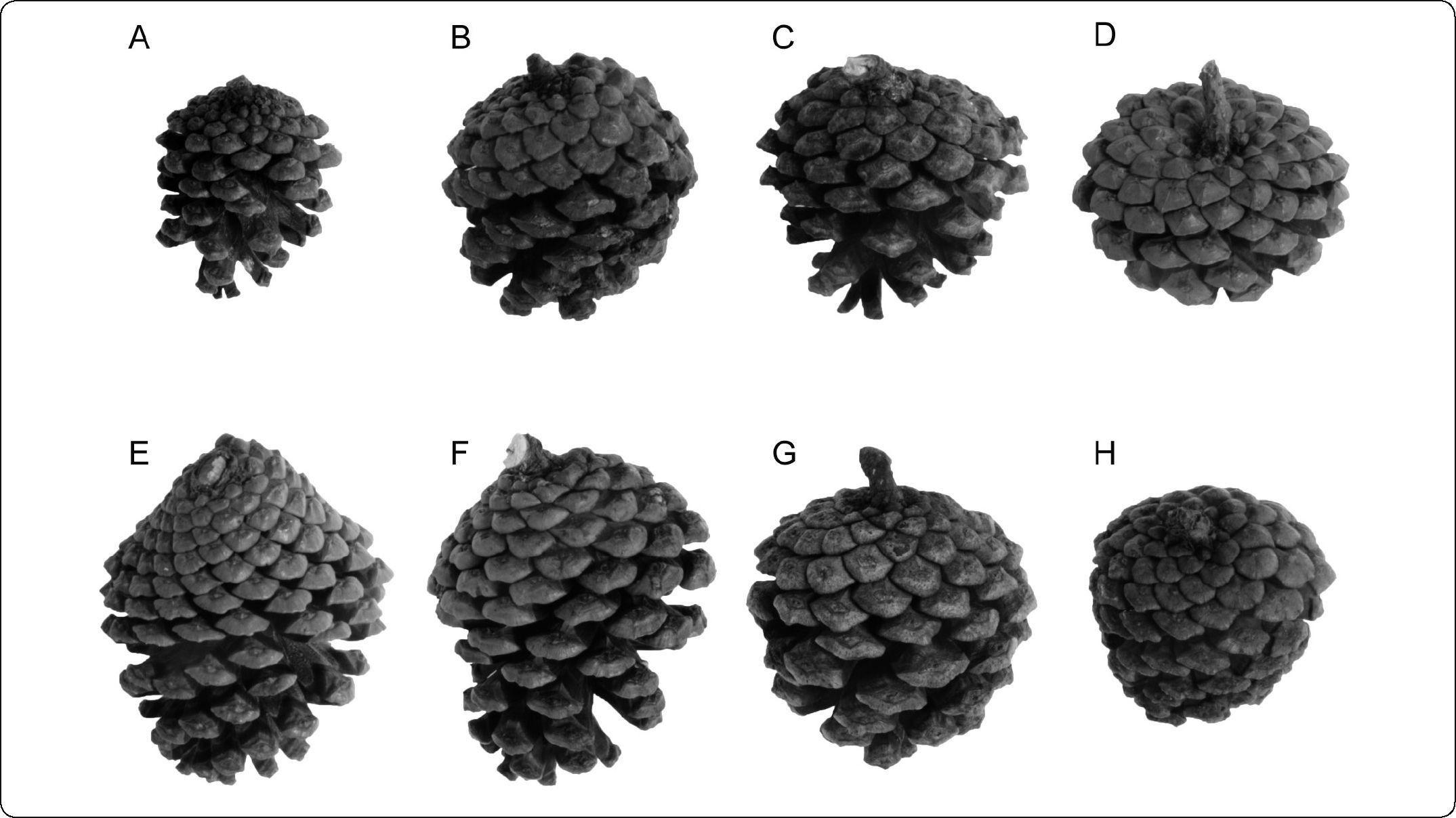
MATERIALS AND METHODSDried specimens were obtained from the Inventario Nacional Forestal and the National Herbarium (MEXU). Ninety individuals were examined morphologically (Figure 1; Appendix 1). Morphological measurements of needles (secondary leaves), fascicle sheaths, seed cones, and peduncles were taken from dried collections. All measurements were made with a ruler and expressed in cm. For needle and fascicle sheath measurements, 15 arbitrarily chosen fascicles were measured. Seed cone and peduncle measurements were based on a single mature cone per individual (Figure 2). Transverse sections were made from the medial part of leaves, fixed in formaldehyde-acetic acid-alcohol (FAA), and examined with an Olympus stereomicroscope. The numbers of stomatal lines on the abaxial and adaxial leaf surfaces were counted, and observations were made of the dermal tissues, mesophyll, and endodermis.
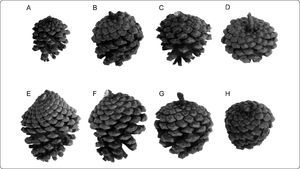
Morphological diversity of seed cones for Pinus subsection Australes in Guerrero and Oaxaca. A. Pinus herrerae González & Martínez 770. B. Pinus lawsonii Soto Núñez et al., 5307. C. Pinus leiophylla Reyes 1989. D. Pinus oocarpa Yescas de los Ángeles 75712. E. Pinus patula var. patula Bautista Martínez 73310. F. Pinus patula var. longipedunculata Trejo 3053. G. Pinus pringlei Rico 489. H. Pinus teocote Román 335. Photographs by Carmen Loyola.
Statistical analyses were performed in R ver. 3.0333. Nine variables were analyzed in 83 individuals for which we could obtain a complete set of measurements: sheath length, number of abaxial stomata, number of adaxial stomata, needle length, needle number, cone length, cone width, cone peduncle length, and cone peduncle width. Four individuals with sessile cones were assigned a peduncle length of 0.01 to avoid using zeros. One-way ANOVAs were performed to test for significant differences among species for each of the nine variables. When significant outcomes were obtained, the Tukey method was used as a post hoc test to identify which species had significant differences between their means. Principal components analysis (PCA) was used to explore the relative importance of the leaf and cone variables for explaining overall variance in the data. PCA was run on the correlation matrix resulting from the log-transformed variables, with their means centered and rescaled.
Leaf or seed megagametophyte tissue was pulverized with a TissueLyser (Qiagen) and genomic DNA was extracted from leaves with the CTAB method34 or from seed megagametophyte tissue using a Wizard Genomic DNA Purification Kit (Promega). Extractions were eluted in buffer (rehydration solution; Promega) and stored at -20°C. The presence of genomic DNA was confirmed by electrophoresis in agarose gel stained with GelRed (Biotium, Inc.).
The first plastid genome, or plastome, reported for a pine was that of P. thunbergii Parl.; it was 119,707 b.p. and composed of 61 protein coding genes, 4, ribosomal RNA genes, 32 tRNA genes, and numerous introns, and intergenic spacers35. Variable sites are not distributed equally throughout the genome. In addition to differences between protein and RNA coding regions and introns and intergenic spacers noncoding regions, there is variation in substitution rates among the different coding regions. The two most variable exons in pine plastomes are called ycf1 and ycf2; both have an elevated nonsynonymous substitution rate and a considerable number of indels, resulting in changes in the amino acid sequences of the proteins that they encode36.
We evaluated variation in an 840 b.p. fragment of ycf1 in Pinus subsection Australes. This fragment represents 13% of the total gene length with respect to the P. thunbergii reference plastome annotation. DNA amplification was carried out using the polymerase chain reaction (PCR). The final concentrations for the PCR were as follows: 1X Buffer, 1.4mM of MgCl2, 0.2mM of each dNTP, 1μM of forward and reverse primers, and 0.625 U of recombinant Taq polymerase. Reactions were carried out as follows: 94°C, 3min; 35 ×(94°C, 1min; 50°C, 1min; 72°C, 2min); 72°C, 5min; 4°C, 5min. The primers used were Pt96887F (5’- tcatttcgaatctttcggattt-3’) and Pt97833R (5’-taccagaatcggacgtgtca-3’)18. The PCR products were sent to the University of Washington High Throughput Genomic Center (Seattle, Washington) for purification and sequencing. The same primers used for PCR were used for sequencing.
Sequence reads were assembled and edited in Geneious ver. 537. Sequences were aligned manually in BioEdit ver. 7.1.938. A haplotype network was inferred using statistical parsimony at a 95% confidence level with TCS ver. 1.2139. Nucleotide diversity (π), the average number of nucleotide differences per site, was also estimated with DnaSP ver. 540.
RESULTSMorphology and leaf anatomyPinus leiophylla Schiede ex Schltdl. & Cham. is the only species of this group in the study area with deciduous fascicle sheaths, a character that is otherwise almost exclusive to soft pines (Pinus subgenus Strobus). Pinus lawsonii Roezl ex Gordon has nondecurrent bracts on its branches and cone peduncle, compared to decurrent bracts in P. pringlei Shaw. Our measurements of needle lengths coincided with previous works6,12,32. The number of leaves per fascicle for P. leiophylla was reported to vary from (2–)3–5 throughout its natural range6,12,13; however, we only found needles in fascicles of 5 or very infrequently 4. The number of leaves per fascicle for P. teocote Schiede ex Schltdl. & Cham. was reported by the same authors to vary from 2–5; however, we found only 3 or very occasionally 4 leaves per fascicle.
An ANOVA F-test indicated that statistical differences existed among species means for all nine morphological variables evaluated (p < 0.001; results not shown). For example, mean fascicle sheath lengths were significantly different (p < 0.05) for 16 of 28 species pairs, mean numbers of abaxial stomata were significantly different for 17 of 28 species pairs, mean needle length was significantly different for 18 of 28 species pairs, and mean cone length was significantly different for 14 of 28 species pairs.
After log transformation, the pairs of variables with the highest correlations were sheath length versus needle length (r=0.649) and the number of abaxial versus dorsal stomatal rows (r=0.844). For the PCAs the cumulative variance for the first, second, and third components were 40.6, 59.7, and 74.5%, respectively. Seven components were needed to explain >95% of the variance. The variables with the highest absolute loading values for the first component were needle length (0.397), number of abaxial stomata (0.390), and number of adaxial stomata (0.386). Needle number was the only variable with a negative value (-0.19). The variables with the highest absolute loading values for the second component were cone peduncle length (0.515), needle number (0.486), and peduncle width (0.334). The variable with the highest negative value for the second component was number of abaxial stomata (-0.312).
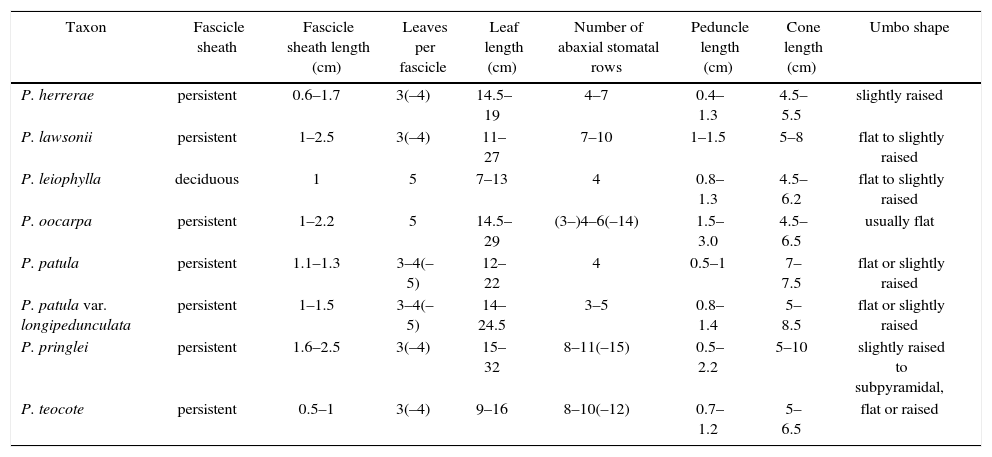
The results of the statistical analyses coincided with our observation that the most useful characters for identifying species of Pinus subsection Australes in Guerrero and Oaxaca were length of the fascicle sheath and needles, the number of rows of stomatal lines, and seed cone peduncle length; qualitative variables such as persistence of fascicle sheaths and several aspects of the seed cone scale were also useful (Table I). Variation in the length or width of the seed cones overlapped greatly among species. Pinus herrerae Martínez had the smallest cones of all species, measuring 4.5–5.5cm in length. The shape of the apophysis and umbo of the cone scale was useful for distinguishing P. oocarpa Schiede, P. pringlei, P. lawsonii, and P. herrerae6,13.
Morphological characters of Pinus subsection Australes species in Guerrero and Oaxaca.
| Taxon | Fascicle sheath | Fascicle sheath length (cm) | Leaves per fascicle | Leaf length (cm) | Number of abaxial stomatal rows | Peduncle length (cm) | Cone length (cm) | Umbo shape |
|---|---|---|---|---|---|---|---|---|
| P. herrerae | persistent | 0.6–1.7 | 3(–4) | 14.5–19 | 4–7 | 0.4–1.3 | 4.5–5.5 | slightly raised |
| P. lawsonii | persistent | 1–2.5 | 3(–4) | 11–27 | 7–10 | 1–1.5 | 5–8 | flat to slightly raised |
| P. leiophylla | deciduous | 1 | 5 | 7–13 | 4 | 0.8–1.3 | 4.5–6.2 | flat to slightly raised |
| P. oocarpa | persistent | 1–2.2 | 5 | 14.5–29 | (3–)4–6(–14) | 1.5–3.0 | 4.5–6.5 | usually flat |
| P. patula | persistent | 1.1–1.3 | 3–4(–5) | 12–22 | 4 | 0.5–1 | 7–7.5 | flat or slightly raised |
| P. patula var. longipedunculata | persistent | 1–1.5 | 3–4(–5) | 14–24.5 | 3–5 | 0.8–1.4 | 5–8.5 | flat or slightly raised |
| P. pringlei | persistent | 1.6–2.5 | 3(–4) | 15–32 | 8–11(–15) | 0.5–2.2 | 5–10 | slightly raised to subpyramidal, |
| P. teocote | persistent | 0.5–1 | 3(–4) | 9–16 | 8–10(–12) | 0.7–1.2 | 5–6.5 | flat or raised |
The anatomical characters of secondary leaves that were compared were the thickness of the hypodermal cell walls, presence or absence of hypodermal cell intrusions into the mesophyll, and position and number of resin canals in the mesophyll (Table II). Endodermal characters were not considered here. All characters coincided with or were within the range of variation reported in previous studies6,12–14.
Leaf anatomical characters of Pinus subsection Australes species in Guerrero and Oaxaca. Pinus tecunumanii is also included for comparison (see text).
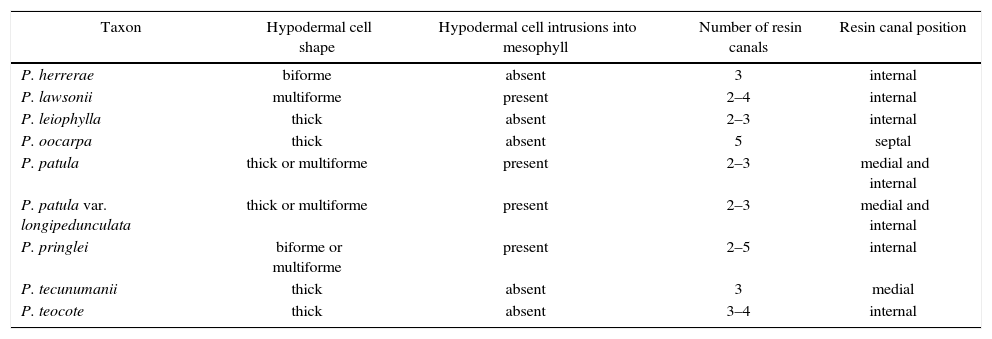
| Taxon | Hypodermal cell shape | Hypodermal cell intrusions into mesophyll | Number of resin canals | Resin canal position |
|---|---|---|---|---|
| P. herrerae | biforme | absent | 3 | internal |
| P. lawsonii | multiforme | present | 2–4 | internal |
| P. leiophylla | thick | absent | 2–3 | internal |
| P. oocarpa | thick | absent | 5 | septal |
| P. patula | thick or multiforme | present | 2–3 | medial and internal |
| P. patula var. longipedunculata | thick or multiforme | present | 2–3 | medial and internal |
| P. pringlei | biforme or multiforme | present | 2–5 | internal |
| P. tecunumanii | thick | absent | 3 | medial |
| P. teocote | thick | absent | 3–4 | internal |
The thickness of hypodermal cell walls was useful for distinguishing P. lawsonii and P. teocote. Intrusions of hypodermal cells into the mesophyll were observed in some transverse sections of P. lawsonii, but never in P. teocote. The position of resin canals was less variable in these two species than has been reported in studies that take into account their entire geographical distribution6,12,13; resin canals were in an internal position (in contact with the endodermis) for both species. Resin canals in a medial position (not in contact with hypodermis or endodermis) were rare. Only P. oocarpa had resin canals in a septal position (in contact with both the endodermis and the hypodermis). Leaf anatomical characters were also useful for confirming morphological identifications of P. patula; none of these individuals agreed in leaf anatomy (e.g., none had five resin canals) with the morphologically similar P. tecunumanii, a species reported for Oaxaca by Farjon & Styles6 but subsequently excluded30. The number of resin canals in the mesophyll was typically 3 for all species except P. oocarpa (5) and to a lesser extent, P. pringlei (2–5).
Identification key for Pinus subsection Australes in Guerrero and Oaxaca- 1.
Fascicle sheaths deciduous, needles in fascicles of 5, 7–13cm long. .. P. leiophylla
1’. Fascicle sheaths persistent, 11–32cm long, needles in fascicles of 3–5. .. 2
- 2.
Needles in fascicles of 4–5, 14.5–29cm long, cone peduncle 1.5–3cm long, seed cones ovoid to subglobose. .. P. oocarpa
2’. Needles in fascicles of 3–4(–5), cone peduncle < 1.5cm long, seed cones ovoid-attenuate. .. 3
- 3.
Needles drooping to pendent, in fascicles of 3–5, 12–22cm long, 4 rows of stomata on abaxial face, 4–6 adaxial, cone sessile or with peduncle ≤ 1cm long. .. P. patula
3’. Needles spreading or erect, in fascicles of 3–4. .. 4
- 4.
Bracts non-decurrent, needles 11–27cm long. .. P. lawsonii
4’. Bracts decurrent. .. 5
- 5.
Prickles on cone scale umbo persistent
- 6.
Needles with 3–5 rows of stomata on abaxial face, 4–6 adaxial, cone peduncle 0.8–1.4cm long × 0.4–0.8cm wide. .. P. patula var. longipedunculata
6’. Needles with 8–11 abaxial rows of stomata on abaxial face, 8–12 adaxial, cone peduncle 0.5–2.2cm long × 0.8–1.3cm wide. .. P. pringlei
5’. Prickles on cone scale umbo deciduous. .. 7
- 7.
Needle length 14.5–19cm, 4–7 rows of stomata on abaxial face, 4–8 adaxial. .. P. herrerae
7’. Needle length 9–16cm, 8–10(–12) rows of stomata on abaxial face, 10 adaxial. .. P. teocote
- 6.
- 5.
- 4.
- 3.
- 2.
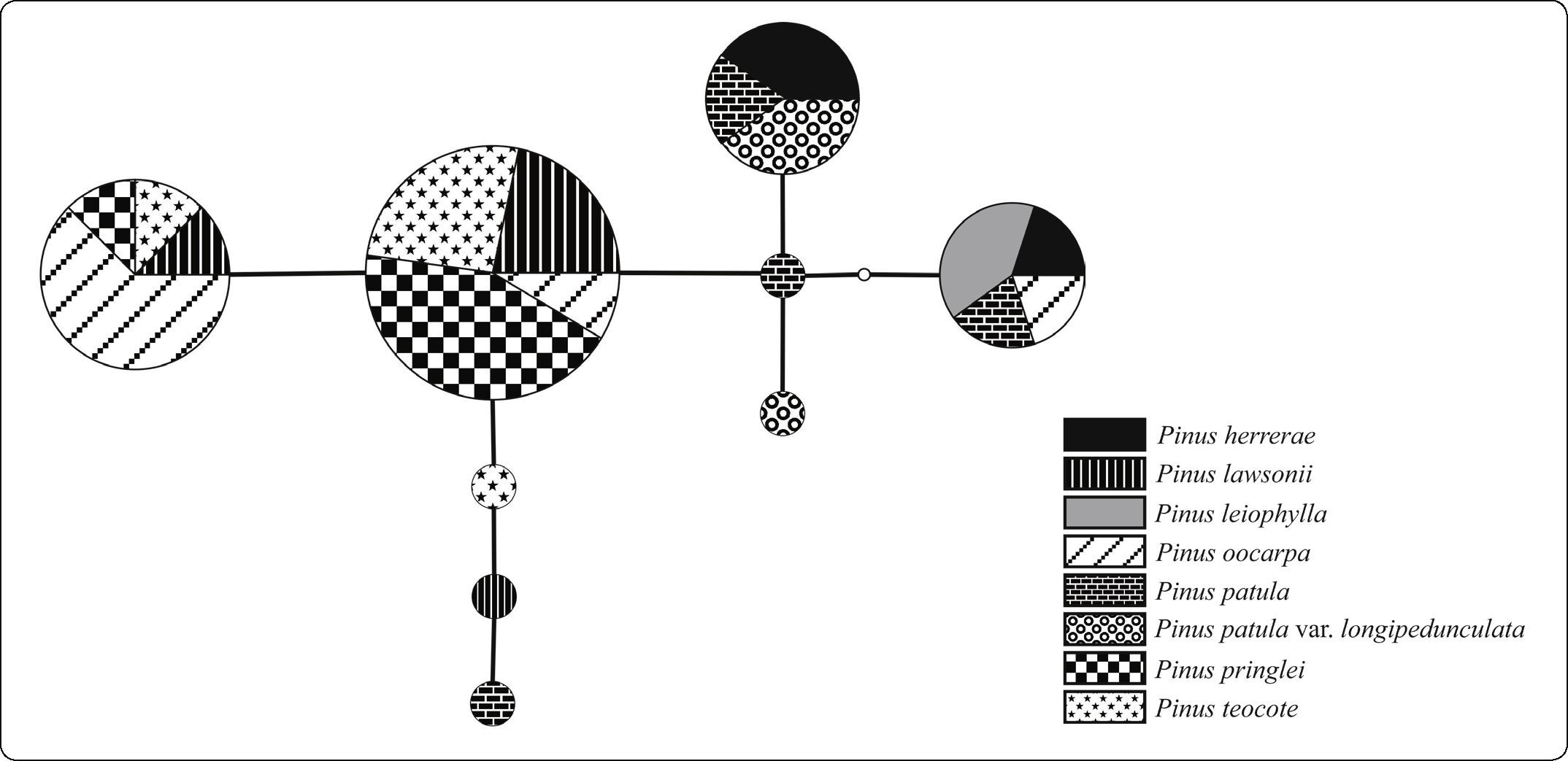
The complete plastid DNA sequence matrix of 49 individuals had a total of seven variable sites; six of the site variants were present in more than one individual (parsimony informative). Nucleotide diversity (π) for all species considered together was 0.00191. The haplotype diversity (Hd) was 0.732. The seven variable sites were distributed among nine different haplotypes, five of which were shared by two or more species (Table III).
Plastid DNA haplotype variation in fragment ycf1 (sequence of primer 5’-3’) and their distribution in the pine species sampled.
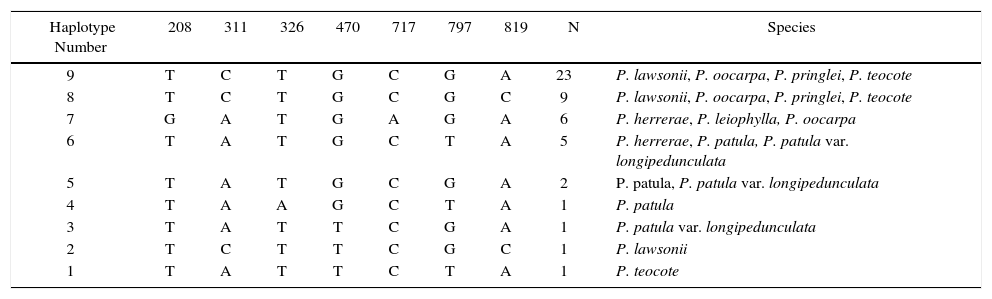
| Haplotype Number | 208 | 311 | 326 | 470 | 717 | 797 | 819 | N | Species |
|---|---|---|---|---|---|---|---|---|---|
| 9 | T | C | T | G | C | G | A | 23 | P. lawsonii, P. oocarpa, P. pringlei, P. teocote |
| 8 | T | C | T | G | C | G | C | 9 | P. lawsonii, P. oocarpa, P. pringlei, P. teocote |
| 7 | G | A | T | G | A | G | A | 6 | P. herrerae, P. leiophylla, P. oocarpa |
| 6 | T | A | T | G | C | T | A | 5 | P. herrerae, P. patula, P. patula var. longipedunculata |
| 5 | T | A | T | G | C | G | A | 2 | P. patula, P. patula var. longipedunculata |
| 4 | T | A | A | G | C | T | A | 1 | P. patula |
| 3 | T | A | T | T | C | G | A | 1 | P. patula var. longipedunculata |
| 2 | T | C | T | T | C | G | C | 1 | P. lawsonii |
| 1 | T | A | T | T | C | T | A | 1 | P. teocote |
The sequences of all the individuals were used to construct a haplotype network (Figure 3). Haplotype 9 occurred at the highest frequency (n = 23) and was shared by the four species P. lawsonii, P. oocarpa, P. pringlei, and P. teocote. It was near the center of the network, with three connections to other haplotypes. The same four species also shared the second most common haplotype (haplotype 8; n = 9). Only one of these four species, P. oocarpa, shared a haplotype with other species in the area, haplotype 7 (n = 6) with P. leiophylla and P. herrerae. In contrast, the two varieties of P. patula only shared haplotypes with each other and with P. herrerae; Haplotype 6, the fourth most frequent in the network (n = 5), was shared among these three taxa. Haplotype 5 (n = 2) was made up of sequences from the two varieties of P. patula; it had the most connections (four) to other haplotypes in the network. The remaining four haplotypes, found in P. lawsonii, P. teocote, P. patula, and P. patula var. longipedunculata, each only occurred once.
DISCUSSIONMorphology is central for the study of taxonomy. It permits the documentation of taxonomic limits, geographic distribution, and phenotypic variation. It is often also useful for the detection of hybrids. In pines, a diversity of leaf types, notably bracts along the branches, fascicle sheaths, and secondary needles, are useful for identifying species, as are seed cone size and shape, length of the peduncle, and morphology of the cone scale apex. Needle anatomy has long been used to delimit or identify species of pines6,11,12. Careful study of morphology and needle anatomy has the potential of identifying putative natural hybrids. The distribution of the species studied here has been described previously31,32, but finer scale information is still lacking. Further field exploration and herbarium work are needed to better establish the geographic distribution of these species in southern Mexico and to capture the full range of morphological and molecular variation of species with ranges that extend beyond Guerrero and Oaxaca.
Forest management practices may be altering the geographic distribution of tree taxa in the area; it is important to establish their natural distributions and determine if interspecific gene flow is being promoted by human activities, particularly the planting of economically important species outside of their natural range of distribution.
The 840 b.p. fragment of plastid DNA characterized in this study offers corroborating evidence of shared plastid DNA haplotypes in North American pines17–23. A previous study that included a 5,425 b.p. plastid DNA alignment for 3–6 individuals per species of Pinus subsection Australes recovered all haplotypes as monophyletic by species for P. herrerae (three individuals), P. leiophylla (four individuals), and P. teocote (three individuals)18. However, in this study we report a P. herrerae individual (and a P. oocarpa individual) with the typical haplotype of P. leiophylla, and two haplotypes in P. teocote that are widespread in other species of Pinus subsection Australes. Further evidence of shared haplotypes, previously reported at a broader geographical scale18, was also found within the study region for P. lawsonii, P. oocarpa, P. pringlei, and P. teocote.
Sharing of plastid haplotypes may be widespread among closely related species of conifers and some other tree genera. The two processes most likely responsible for this phenomenon in pines are introgressive hybridization and incomplete lineage sorting. These processes are probably most widespread in groups of recently diverged species, such as within Pinus subsection Ponderosae and Pinus subsection Australes18, or in young putative species pairs, such as the Mexican white pines, P. ayacahuite and P. strobiformis22. Less plastid haplotype sharing has been reported from other conifer lineages with fewer recently diverged species such as North American pinyon pines41,42. Some recently diverged lineages, such as P. caribaea of Pinus subsection Australes43 also show little evidence of shared haplotypes, possibly due to less co-occurrence with close relatives and smaller effective population sizes. Distinguishing between introgressive hybridization and incomplete lineage sorting can be achieved by comparing the genealogical patterns of plastid DNA with those of nuclear and mitochondrial DNA, both of which are likely to result in different patterns than those found with plastid DNA. Further evidence of hybridization can also be documented through morphological and anatomical studies.
CONCLUSIONSGuerrero and Oaxaca are rich in species of Pinus subsection Australes. They occur both allopatrically and sympatrically throughout the montane regions of the state. Although they can be distinguished morphologically, they follow a more generalized pattern in North American hard pines of sharing plastid DNA lineages as a result of introgressive hybridization or incomplete lineage sorting. Inferring plastid DNA genealogies is of unquestionable value for studying evolution in the group, but it is important to be aware that adequate infraspecific sampling and inclusion of sufficient variable sites are needed for plastid DNA genealogies to be informative with respect to species relationships.
Future studies should incorporate range-wide sampling of species to capture their full range of variation. Sequences from multiple unlinked markers combined with morphometric analysis should permit more accurate detection of interspecific gene flow in the presence of incomplete lineage sorting.
This study formed part of the undergraduate thesis of Alfredo Ortiz-Martínez for the Instituto Tecnológico de Cd. Altamirano. We thank Mauricio Mora for assistance in the herbarium, Carmen Loyola for providing photographs of seed cones, and Jazmín Cortez Sarabia, Erika Oropeza Bruno, Jesús Pascual Orozco, Francisco Zavala Hernández, Lev Jardón Barbolla, and two anonymous reviewers for providing helpful comments on a previous draft of this manuscript. Most of the plant material in this study was provided by the project “Remuestreo del Inventario Nacional Forestal y de Suelos 2009-2013” awarded by the Comisión Nacional Forestal (CONAFOR) to Martin Ricker. We thank the Dirección General de Asuntos de Personal Académico-Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica of the Universidad Nacional Autónoma de México (DGAPA-PAPIIT; IN228209) for financial support for laboratory work, and CONACYT (INFR-2014-1 No. 224743) for supporting the installation of herbarium compactors that have allowed for the storage of the specimens for this study.
Pinus herreraeMartínez
México. Oaxaca. Loma El Ocote, I. Martínez Martínez 71204. Guerrero. Cerro La Mula, E.O. Valencia Santana 71124. Cerro Faisán, M.A. Juárez 72227. Omiltemi, Chilpancingo, J. A. Pérez de la Rosa 132. Camino Paraíso, L. Lozada 182A.
Pinus lawsoniiRoezl ex Gordon
México. Oaxaca. Arroyo Cangarec, O. Rodríguez Santamaría 74754. La Ocotera, O. Rodríguez Santamaría 73947. Arroyo Artemio Reyes, G.R. Izaguirre Yáñez 76960. Barranca Pelona, J.C. Calvillo García 71396. El Zacatón, J. Zúñiga Reyes 74549. Agua Cola, J. Zúñiga 74747. Pino Gordo, O. J. Pérez Martínez 72853. Nanche, R, Boquedano Peralta 76955. Guerrero. Exsuyo, J.C. Calvillo García 71587. La Laguna, J.C. Calvillo García 70214. El Chaude, A.J. Fortoul Velasco 70254.
Pinus leiophyllaSchiede ex Schltdl. & Cham.
México. Oaxaca. La Plaga, R. Baquedano Peralta 72700. Monte de Tesoro, J. Reyes Santiago 883. Miahuatlán, E. Hunn 0096. Santa María del Rosario, A.J. Fortoul Velasco 72252. Ixtlán, A. García-Mendoza 7585. Guerrero. La Guitarra, A.J. Fortoul Velasco 71557. La Culebra, A.J. Fortoul Velasco 71773.
Pinus oocarpaSchiede ex Schltdl.
México. Oaxaca: El Corolón-El Retén, T. Yescas de los Ángeles 75410. Agua Fría, T. Yescas de los Ángeles 75712. Loma del Chifle, T. Yescas de los Ángeles 76958. La Ermita, J.L. Flores Nicanor 71650. La Cascada, E. Martínez V. 73980, Rancho Viejo, T. Yescas de los Ángeles 72018. Guerrero. La Sidra, J.C. Bautista Martínez 71991. Agua de Obispo, J.C. Calvillo García 72424. La Coscolina, O.J. Pérez Martínez 72838. El Ermitaño, J.C. Calvillo García 70448. Tierra Blanca, J.C. Calvillo García 72023.
Pinus patulaSchltdl. & Cham. var.patula
México. Oaxaca. Terracería La Venta, J. Reyes 6493. La Cima, M.I. Alvarado Flores 72489. Lomatico, J.C. Bautista Martínez 73310.
Pinus patulavar.longipedunculataLoock ex Martínez
México. Oaxaca. Aserrado Las Vigas, J. A. Pérez de la Rosa 1849. San Juan Tepeuxila, R. Torres Colín 16168. Santa María Yavesía, I. Trejo 3064.
Pinus pringleiShaw
México. Oaxaca. Nebrón, J. Zúñiga Reyes 71153. La Caoilla, Ojo de Agua, M. I. Alvarado Flores 69610. El Chaneque, J.L. Flores Nicanor 75148. Silacayuapan, R. Luna Martínez 71600. Tres Cruces, J.L. Flores Nicanor 74544. San Isidro El Chiñón, R. Luna Martínez 71813. Buenavista, O. Rodríguez Santamaría 70069. Guerrero. Cajones, J.C. Calvillo García 65501. Cocuitlazola, J.C. Calvillo García 72446. C. Tlacholoya, J.C. Calvillo García 71371. Ejido El Carrazal, Coyuca de Catalán, E. Rodríguez Ibarra 69525. Huexoapa, F. Rosas Aguilar 72658.
Pinus teocoteSchiede ex Schltdl. & Cham.México. Oaxaca. Agua Colorada, I. Martínez Martínez 74372. Cerro Manteca, J.L. Flores Nicanor 75345. Yacuni, M.I. Alvarado Flores 72898. La Ciénaga, J.L. Flores Nicanor 73537, La Joya del Fresno, T. Yescas de los Ángeles 72476. Río Milpas, R. Baquedano Peralta 73534. Barranca del Zopilote, J.C. Calvillo García 71607. San Francisco Higos, R. Lina Martínez 72028. Guerrero. Mesa de la Mujer, A.J. Fortoul Velasco 68303. San Andrés, A.J. Fortoul Velasco72003.