Recent years have witnessed the advent and rapid development of massive sequencing technology, commonly known as Next Generation Sequencing (NGS). This technology allows for rapid, massive and inexpensive sequencing of genome regions or entire genomes, making possible genomic studies of non-model organisms and has seen great progress in metagenomic studies. The promise of this information-rich era is to expand the molecular approach of ecological and evolutionary studies towards urgent issues related with conservation and management of biological diversity in the face of global change. Among the current NGS technologies, there are fundamental differences that impact DNA sequence accuracy, length and range of applications. Key differences among platforms are the procedure for library preparation (when needed) and the sequencing process itself (e.g., pyrosequencing, synthesis). In this review we describe the technical details of commercially available platforms for massive sequencing. We discuss their potential applications for specific biodiversity analyses, from model to non-model organisms, from Single Nucleotide Polymorphism (SNPs) to entire genome analysis and metagenomic approaches of microbial communities, including possible taxonomic, phylogenetic, conservation biology and ecosystem applications of NGS methods in the study of biodiversity. We also provide a to-date estimation of the associated costs for each approach and the computational implications for the analyses of sequences derived from these platforms.
Recientemente se han desarrollado nuevas tecnologías de secuenciación masiva, conocidas como secuenciación de siguiente generación (NGS, por sus siglas en inglés). Estas tecnologías permiten secuenciación rápida, masiva y a bajo costo de regiones genómicas o genomas completos, haciendo posible estudios genómicos de organismos no modelo y estudios metagenómicos. Estas tecnologías prometen expandir las aproximaciones moleculares de estudios ecológicos y evolutivos hacia asuntos relacionados con conservación y manejo de la diversidad biológica ante retos como cambio climático. Entre las plataformas NGS disponibles hay diferencias fundamentales que resultan en diferente precisión en la determinación de las secuencias, así como diferencias en la longitud de las mismas. Algunas diferencias clave entre plataformas son los procedimientos para la preparación de bibliotecas (cuando son necesarias) y el proceso de secuenciación per se (e.g., pirosecuenciación, síntesis). En esta revisión se describen las plataformas comercialmente disponibles para NGS y se discuten sus aplicaciones en estudios de biodiversidad de organismos modelo o no modelo, como son análisis de polimorfismos únicos (SNPs), así como análisis de genomas completos y aproximaciones metagenómicas para el estudio de comunidades microbianas. También revisamos posibles aplicaciones de los métodos NGS para resolver problemas taxonómicos, filogenéticos, de biología de la conservación y de ecosistemas, todos relevantes en el estudio de la biodiversidad. Adicionalmente se presenta una estimación actual de los costos asociados para cada plataforma, así como las implicaciones computacionales para los análisis de secuencias derivadas de estas tecnologías.
In recent years, development of massive sequencing technology has permitted rapid and relatively inexpensive sequencing of large portions or even entire genomes of different organisms. These technologies, in contrast with more traditional sequencing methods, allow sequencing non-model organisms for which limited genetic information is available (Mardis, 2008; Neale and Kremer, 2011; Cahais et al., 2012). Moreover, the field of metagenomics has flourished with the advent of massive sequencing technologies, which broadens the range of questions that can be posed and answered from ecological and evolutionary perspectives in conservation and management of natural resources (Bonilla-Rosso et al., 2008; Eguiarte et al., 2013).
The overarching goal of genomic studies is the understanding of diversity, defined as genetic variation, nucleic acid sequence variation, and the comparison of such variation among organisms (Hedrick, 2000; Eguiarte et al., 2013). This goal can be accomplished in different ways, depending on the scale at which the analysis is conducted (Mardis, 2008; Metzker, 2010; Neale and Kremer, 2011; Zhang et al., 2011). For example, the analysis can be limited to determining the sequence of a region of the genome or the complete genome of the organisms under study, including organelles in the case of eukaryotes. Furthermore, the analysis of sequences can be taken to another level by looking at the physical position of each base in a genetic map, and even further with molecular evolution and population genetics analyses that have seen considerable advances since genomic data became available (Turner and Hahn, 2007; Michel et al., 2010; Yi et al., 2010). As examples of the opportunities that genomics offer to population genetics studies, it has been shown that when comparing different lineages, it is possible to determine the physical arrangement of genes and their evolutionary conservation (e.g., synteny; Mathee et al., 2008), the evolutionary dynamics of species and genomes (e.g., Salmonella; Holt et al., 2008), gene and genome duplications and architecture (Ibarra-Laclette et al., 2013), the evolutionary history of species (phylogenomics; (Delsuc et al., 2005) and the genetic targets of selection and the genetic basis of adaptation (Yi et al., 2010).
Moreover, genomic studies offer great promise in advancing our understanding of complex processes associated with gene expression and gene interactions (i.e., transcriptomics, proteomics, epigenomics, developmental genetics, epistasis, pleiotropy, etc.), as well as to take a better grip into the genetic basis of phenotype and the complex relationship genotype-phenotype-environment (Mardis, 2007; Mardis, 2008; Neale and Kremer, 2011). Finally, the new field of metagenomics, in which the goal is the sequencing of all genomes from an environmental sample, has been particularly benefited from massive sequencing (Thomas et al., 2012).
Given the great insight that genomic information offers to understanding biodiversity, we consider very important to revise currently available tools that researchers in natural sciences can use if they decide to pursue a genomic approach in their studies. In this review, we describe technical details of the commercially available platforms for massive sequencing, as well as their associated costs and computational demands. We also discuss the potential applications of these platforms for specific biodiversity analyses, either ecological or purely evolutionary, from model to non-model organisms, from Single Nucleotide Polymorphism (SNPs) across the genome of regions of unknown identity or specific genes, to entire genome analysis and metagenomic approaches of microbial communities.
Sequencing basics: from Sanger to next generation sequencing (NGS)For almost 3 decades, sequencing efforts were carried out by the classic Sanger method (Sanger et al., 1977), which is still the most used approach for routine molecular analyses. This method allows determination of nucleotide sequence of DNA fragments in the range of 1000 base pairs (bp), which approximates the length of an average gene. Sanger sequencing method signified a great improvement with respect to the Maxam-Gilbert (1977) method, which requires 32P as radioactive marker with the inconvenience of the relatively long decay rate. Moreover, and in contrast to Sanger or even some next generation methods for sequencing, the Maxam-Gilbert method is not based in the copy or synthesis of a template DNA strand, but in the inference of the sequence as a result of chemical alterations or enzymatic restrictions of the sequence of interest (Maxam and Gilbert, 1977). The simplicity of Sanger method or dideoxi, based on chain-termination synthesis made possible its popularization and establishment as the standard sequencing method. Nonetheless, Maxam-Gilbert method is still used in some cases to identify epigenetic modifications, such as methylation (Church and Gilbert, 1984; Isola et al., 1999; Ammerpohl et al., 2009).
Optimization and automation of sequencing by Sanger method was possible thanks to its coupling with PCR (polymerase chain reaction) technique (Mullis and Faloona, 1987) to obtain multiple copies of a specific DNA fragment, as well as with the introduction of fluorescently marked dideoxinucleotides instead of the originally radioactively marked ones (Smith et al., 1986). Today, automated sequencing by Sanger method requires the generation of relatively small fragments (1 000 bp), either by selective amplification by PCR of the regions of interest (amplicons), or by physical or chemical fragmentation of entire genomes, a strategy known as shotgun sequencing (Staden, 1979). Once the fragments are generated, these can be sequenced. If the fragments are obtained through amplification of a single homozygous individual, sequencing can proceed through synthesis. In the case of heterozygous or a mixture of individuals (populations) or species (communities, metagenomics), and also in the case of shotgun generated fragments, individualization of the different copies is needed and is usually accomplished by clone library construction (Escalante, 2008).
The shotgun strategy was of great relevance and increased the rate at which genomes were sequenced, becoming the most used approach for genome sequencing for many years (Anderson, 1981; Roach et al., 1995; Adams et al., 2000). In fact, this technology allowed sequencing of the first complete genomes, including the human genome (International Human Genome Sequencing Consortium 2001). However, this massive approach has two main difficulties: i) each sequencing reaction has to be performed separately, which results in a limited number of base pairs obtained per day, and ii) the cost per base pair is relatively high, which at a genomic scale can be prohibitive for most laboratories and research institutions.
As we will describe below, massive sequencing methods couple clone library construction with sequencing in different ways. In other words, one common trait of all platforms of massive sequencing is a 2 level parallelization (Liu et al., 2012): generation and separation of millions of individual fragments, and their subsequent parallel sequencing (Mardis, 2008). It should be noted that some next generation sequencing strategies promise real time sequencing of single molecules, eliminating the clone library construction step (Thompson and Milos, 2011; Carneiro et al., 2012).
Another common element that has been important in the development of massive sequencing technologies is the progressive incorporation of advances in materials science. This allows the use of thin layers, nanopores, nanoscale emulsions or microspheres as physical means to adhere DNA fragments and probes. This miniaturization of the scale of work is a key aspect of the massive parallelization of the processes involved in simultaneously obtaining of the nucleotide sequences of thousands and millions of fragments.
Besides the specific details of clone library construction and sequencing strategies, to date all sequencing platforms have the same basic stages of sample processing. These stages are: i) total DNA or RNA extraction; ii) amplification of specific genomic regions or fragmentation (shotgun) of total DNA. Fragmentation can be done enzymatically or physically (e.g., nebulization, sonication). In case of RNA sequencing, retrotranscription is required to obtain cDNA which in turn is fragmented; iii) selection of fragments by size (usually 200 bp); iv) adaptor sequence addition to the fragment mix; v) library construction (except real time sequencing technologies, and vi) sequencing.
It is important to note that in all cases during the preparation of fragments it is possible to incorporate specific sample-labels to simultaneously sequence multiple samples, a procedure that is technically known as multiplexing or barcoding. The uniqueness of the next generation sequencing methods resides in the fact that addition of a new nucleotide generates some type of physicochemical change (i.e., light emission, voltage change, fluorescence) that can be detected by an ultrasensitive device coupled to the sequencing system itself.
In the following section we will describe the strategies of library construction and sequencing in detail for the currently available sequencing platforms.
Next generation sequencing methods or massive parallel sequencingCoverage is a central concept to the description of capacities, limitations and error rates of massive sequencing methods. It refers to the number of times that a specific nucleotide is read during the sequencing process (DePristo et al., 2011). Coverage of 30X means that, on average each base has been read 30 times. However, coverage if not uniform across regions, with regions having low or null coverage and regions being well covered. This variation in coverage across regions follows a certain distribution, sometimes called coverage depth or depth histogram (http://www.illumina.com/truseq/quality_101/coverage/coverage_distribution.ilmn).
Higher coverage guaranties that almost all regions have been sequenced at least once. However, it increases laboratory and bioinformatics costs while increasing certainty of base assignment. Giventhat DNA fragmentation generates a collection of pieces of different size and with some level of overlap among them, a single nucleotide of the same sequence will be represented many times during the amplification and sequencing processes. In general terms, platforms with longer reads (more bp) tend to generate less copies of the same nucleotide, thus have less coverage. On the other end, platforms with smaller fragments tend to deliver more copies of the same nucleotide, improving coverage but requiring more computational resources for adequate sorting and assembly.
The level of coverage gains relevance because of the elevated error rate of the available methods of massive sequencing in comparison to Sanger sequencing (Table 1). More reads give confidence to the base assignment during the reading of sequences and will also simplify the assembly and sorting during the bioinformatics processing. Moreover, biases during library construction may result in an overrepresentation (more coverage) of particular fragments or amplicons, which will alter considerably the results and their interpretation. These biases can have technical (procedural) causes, may be produced by intrinsic sample characteristics (CG content, DNA extraction biases) or can be associated with platform-inherent and poorly understood causes.
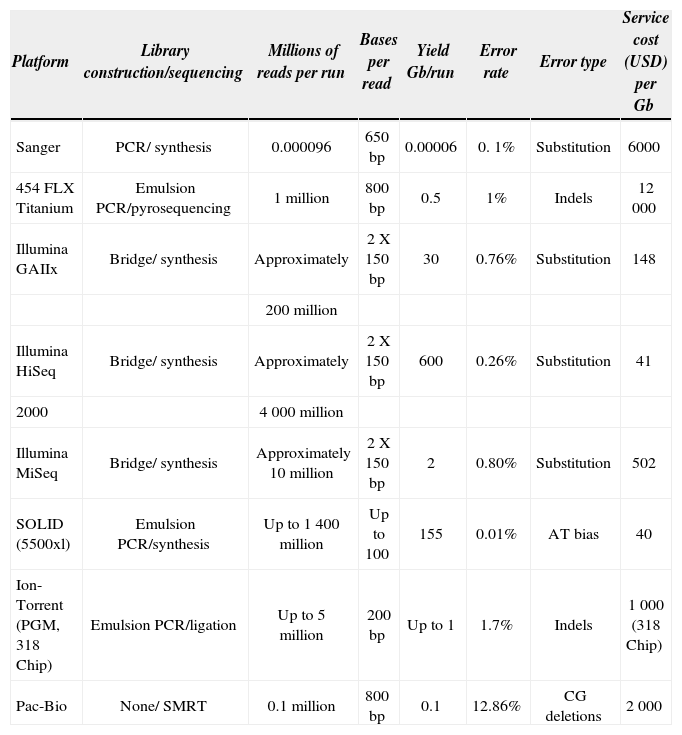
Next generation sequencing platform traits (modified from Glenn, 2011 and Quail et al., 2012)
| Platform | Library construction/sequencing | Millions of reads per run | Bases per read | Yield Gb/run | Error rate | Error type | Service cost (USD) per Gb |
|---|---|---|---|---|---|---|---|
| Sanger | PCR/ synthesis | 0.000096 | 650bp | 0.00006 | 0. 1% | Substitution | 6000 |
| 454 FLX Titanium | Emulsion PCR/pyrosequencing | 1million | 800bp | 0.5 | 1% | Indels | 12 000 |
| Illumina GAIIx | Bridge/ synthesis | Approximately | 2 X 150bp | 30 | 0.76% | Substitution | 148 |
| 200million | |||||||
| Illumina HiSeq | Bridge/ synthesis | Approximately | 2 X 150bp | 600 | 0.26% | Substitution | 41 |
| 2000 | 4 000million | ||||||
| Illumina MiSeq | Bridge/ synthesis | Approximately 10million | 2 X 150bp | 2 | 0.80% | Substitution | 502 |
| SOLID (5500xl) | Emulsion PCR/synthesis | Up to 1 400million | Up to 100 | 155 | 0.01% | AT bias | 40 |
| Ion-Torrent (PGM, 318 Chip) | Emulsion PCR/ligation | Up to 5million | 200 bp | Up to 1 | 1.7% | Indels | 1 000 (318 Chip) |
| Pac-Bio | None/ SMRT | 0.1million | 800bp | 0.1 | 12.86% | CG deletions | 2 000 |
Among the currently available platforms for massive sequencing, the most used are the second generation platforms, which include 454, Illumina, SOLID and Ion-Torrent. We will describe each one in terms of their procedures for library construction and sequencing strategies.
Roche / 454 GLS FLX Titanium Pyrosequencer. This platform is commonly referred to as pyrosequencer. The fragments to be sequenced, as we previously mentioned, have to be selected by size (400-600 bp), and then processed to ligate the adaptors necessary for library construction. The procedure for library construction relies on PCR amplification of each DNA fragment in the sample. Each amplification reaction happens inside a micelle that contains a single DNA molecule, a nanosphere or bead, a polymerase molecule and the rest of the components necessary for the reaction to proceed (dNTPs, buffer, MgCl2, primers). This process is known as emulsion PCR.
Each bead has oligonucleotides or probes attached to its surface, the sequence of these probes is complementary to the fragment adaptors. It is important to quantify with precision the components of the PCR reaction, because each micelle has to contain only 1 DNA molecule in order to create a clonal amplification attached to each bead. After library construction, the resulting emulsion, which contains the beads (each bead is a clone), is deposited in a picotiter plate (PTP). The PTP allows only 1 bead in each of the hundred thousand wells that are individually monitored during the pyrosequencing process. Data for an individual DNA sequence will be produced in each well.
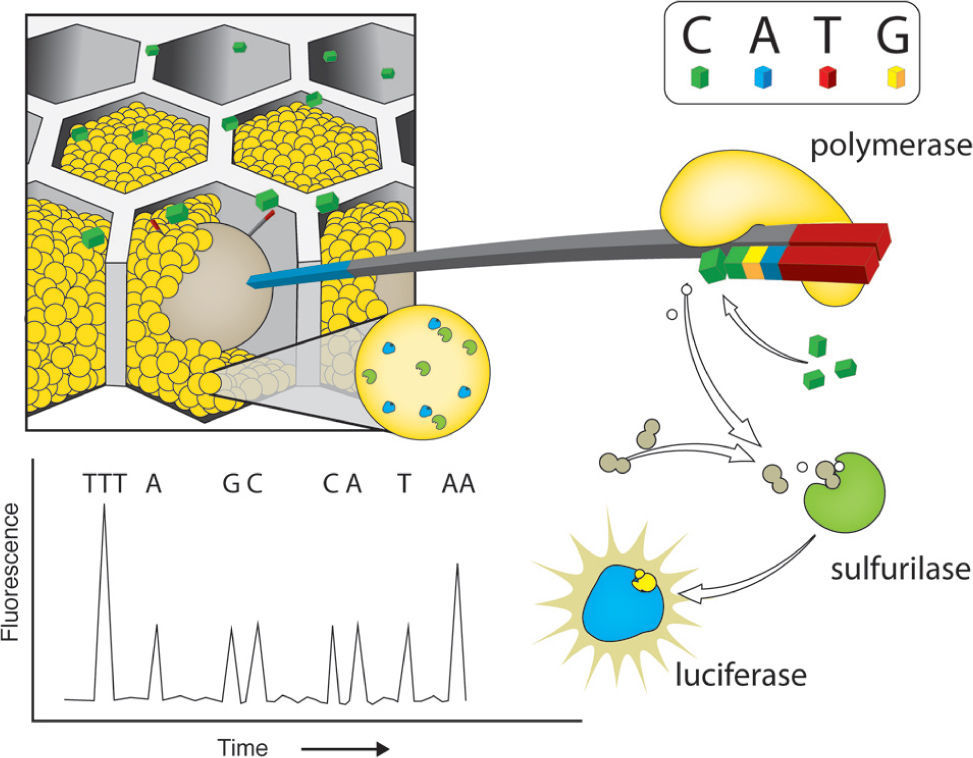
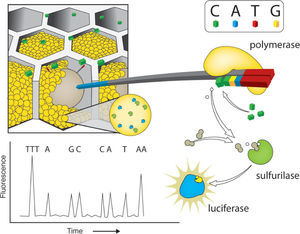
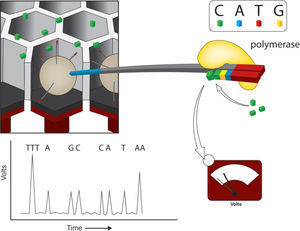
Pyrosequencing reactions consist on the synthesis of the complementary strand of the DNA molecules attached to the beads. This reaction is coupled with the activity of enzyme containing beads that are also added to the PTP and surround the DNA beads. Enzymatic activity of luciferase and sulfurilase catalyze the downstream pyrosequencing reaction steps. The PTP serves as a flow cell into which single nucleotide solution is added 1 at a time. The addition of nucleotides results in light emission and no enzymatic termination. The PTP is coupled with a high-resolution camera (CCD) that takes pictures every nucleotide round. Nucleotide addition is not followed by termination and thus the first nucleotides of the adaptor serve as a light emission calibration equivalent to 1 nucleotide, allowing the calculation of the number of nucleotides added in a single round of synthesis (Fig. 1). However, the calibration for long stretches of a single nucleotide has its limits, and it is usually inaccurate when > 6 bases of the same nucleotide are added in a row (Mardis, 2008). Although in the last years the length of reads obtained by pyrosequencing has increased considerably (Shokralla et al., 2012), the coverage for this approach is still the lowest for the currently available platforms, ranging from 7-10X, depending on genome size (i.e. Barbazuk et al., 2007; Wheeler et al., 2008; Vera et al., 2008).
Pyrosequencing using Roche/454 Titanium platform. After loading the DNA-amplified beads (libraries) into individual Pico Titer Plate (PTP) wells, other type of beads, coupled with luciferase and sulphurilase, are added. The figure shows just one type of 2'-deoxyribonucleoside triphosphate (dNTP) - cytosine - that flows through the wells. Once the polymerase adds one nucleotide, a sulfurilase-luciferase reaction occurs, emitting light. A fiber optic slide is attached to a microfluidics camera allowing the reagents to reach the wells packed with beads. Underneath the fiber optic slide there is a direct connection to a high-resolution camera (charge coupled device or CCD), which allows detection of light emitted by each PTP when a pyrosequencing reaction occurs. Modified from Metzker (2010).
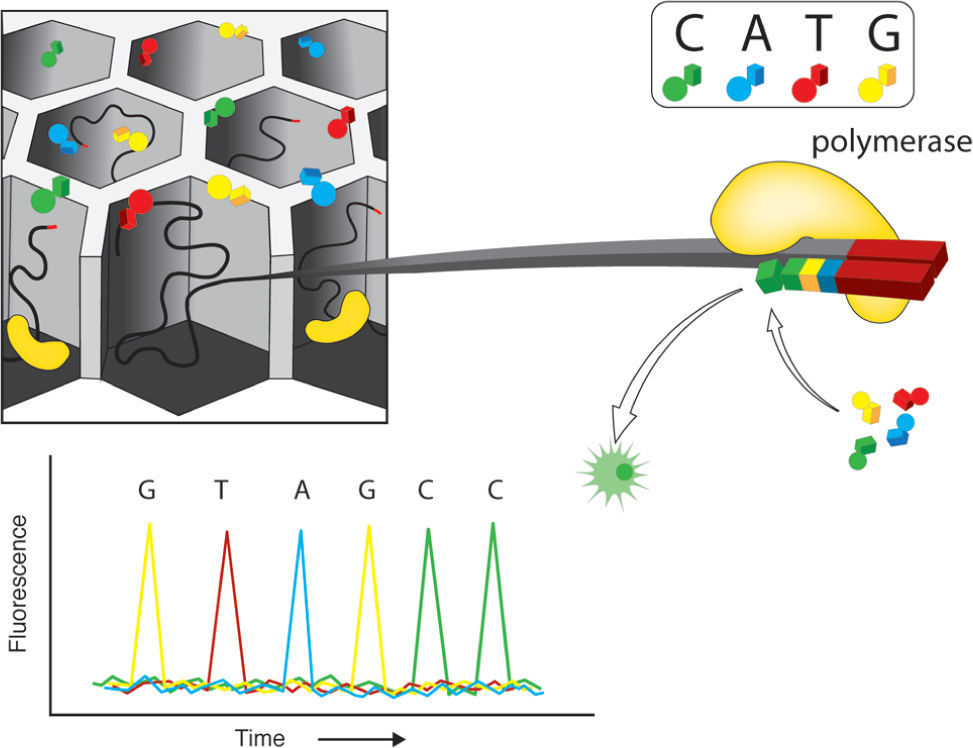
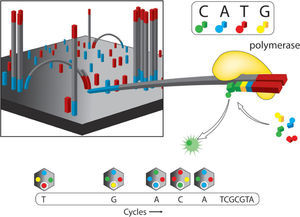
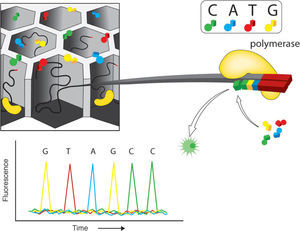
Illumina genome analyzer (GAIIx). This is one of the most praised massive sequencing platforms, due to its high quality sequences and great coverage (millions of simultaneously sequenced fragments). The procedure for library construction is also based on synthesis of complementary strands of DNA through PCR. However, the specifics of such synthesis have notable differences with the emulsion PCR used by 454. As most current massive sequencing protocols, DNA fragmentation and size selection are necessary steps prior to library construction. Once fragments of a desired size are selected, platform-specific adaptors are added which allows attachment to the sequencing matrix, a flow-cell device that will support bridge amplification and sequencing per se. On this matrix, each attached fragment is amplified producing multiple and identical DNA copies in a cluster (Fig. 2). Illumina's matrix has 8 separate channels (or lines) in each of which a library of clusters can be created. Each cluster is sequenced by synthesis, in which all 4 nucleotides are added simultaneously to the flow device, along with DNA polymerase for addition into the oligo-primed fragments that form the clusters. For this sequencing approach, the addition of a nucleotide blocks subsequent incorporation of nucleotides, interrupting the synthesis, and releases a fluorescent signal that is unique for each type of nucleotide. Imaging of the fluorescent signal follows the incorporation. After imaging, the blocking group is removed and leaves DNA strands ready for the next nucleotide incorporation by DNA polymerase. This series of steps continues for a specific number of rounds allowing read-lengths of 60-150 bases (Glenn, 2011).
Illumina. Solid phase amplification (upper left). The initial step is the coupling of oligonucleotides (priming) and the extension of single-strand molecules. The next step is bridge amplification of the immobilized template with the adjacent primer to form clusters. Four fluorescent color reversible termination (upper right). This platform uses a terminator and fluorescent label that differs for each base type. After the addition of a new base, a picture is taken and fluorescence removed leaving the hydroxyl group free for a new addition (bottom). As an example, the bottom part of the figure shows 4 color images that represent the sequence data from one template. Modified from Metzker (2010).
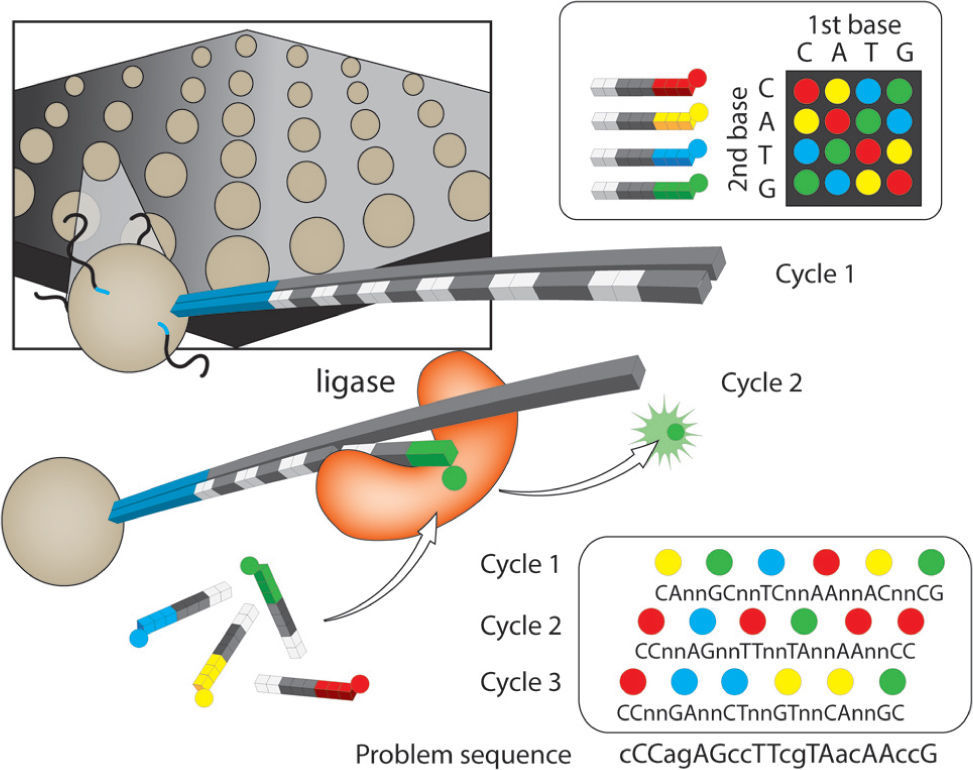
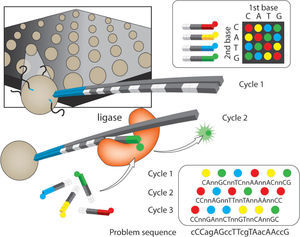
Applied biosystems SOLiD™ sequencer. Sequencing by oligo ligation detection (SOLiD). This platform has the best quality of sequences and the smallest error rate (Table 1), as a result of the ligation-based approach. The library construction occurs through an emulsion PCR with small magnetic beads (similar to 454/Roche). After completion of library construction, the resulting beads are attached covalently to a flow-cell glass slide. Compared to other platforms, SOLiD has a major difference in that the approach taken for sequencing the amplified fragments, uses DNA ligase as illustrated in figure 3. The slide with attached beads is exposed to cyclic enzymatic reactions. During the first cycle a primer sequence is incorporated, that is complementary to the adaptor ligated to the fragments as well as a ligase and 4 fluorescently labeled 2-base encoded probes. Non-ligated probes are then washed away, followed by the imaging of the released fluorescence that identifies the ligated probe (Landergren et al., 1988). The cycle is repeated to remove the fluorescent dye and regenerate the 5'-PO4 groups for 10 subsequent ligation cycles (Fig. 3). A second ligation round is performed with a “n-1” primer, which resets the interrogated sequence one base downstream, and then the 10 cycle ligation proceeds.
SOLiD. Four color sequencing by ligation. After annealing of a universal primer, a library of 1-2-probes is added. In contrast with sequencing by synthesis, sequencing by ligation involves ligation of the probe to a universal primer. The ligation event releases fluorescence and after imaging, ligated probes are chemically removed to generate a 5'-PO4 group. The cycle is repeated 9 more times, after which the primer is removed. Upon primer removal, another primer is added but with a one base pair shift towards the 3-'end. Four base pair shifts are completed with the corresponding 10 ligation cycles. The 1-2-probes are designed to interrogate the first (x) and second (y) positions adjacent to the hybridized primer, such that the 16 nucleotides are encoded by 4 dyes. Two-base color scheme. There are 4 dinucleotide sequences associated with a color (e.g. AA, CC, GG, TT are coded with a blue dye). Each template or sequence is interrogated twice and compiled in a color space. The readings of the color space are aligned against a reference to decode the DNA sequence. An example of the double interrogation is illustrated for three out of the 5 cycles that complete the reading (lower left). Upper case letters denote double interrogated bases, lower case letters are bases interrogated once. If 5 cycles were included, all bases would have been interrogated twice. Modified from Metzker (2010).
Four more rounds of ligation cycles are performed with progressively “n-1” primers. Color calls from the 5-ligation rounds are then ordered into a linear sequence (the color space) and compared to a linear sequence to decode DNA problem sequence. The read length of SOLiD was initially 35bp with a final output of 3Gb per run (Liu et al., 2012). These numbers have increased up to 75bp per read and 10Gb per run (Shokralla et al., 2012), with high accuracy in the assignment of bases due to the 2-base sequencing method (accuracy of 99.85% after filtering; Liu et al., 2012; Table 1).
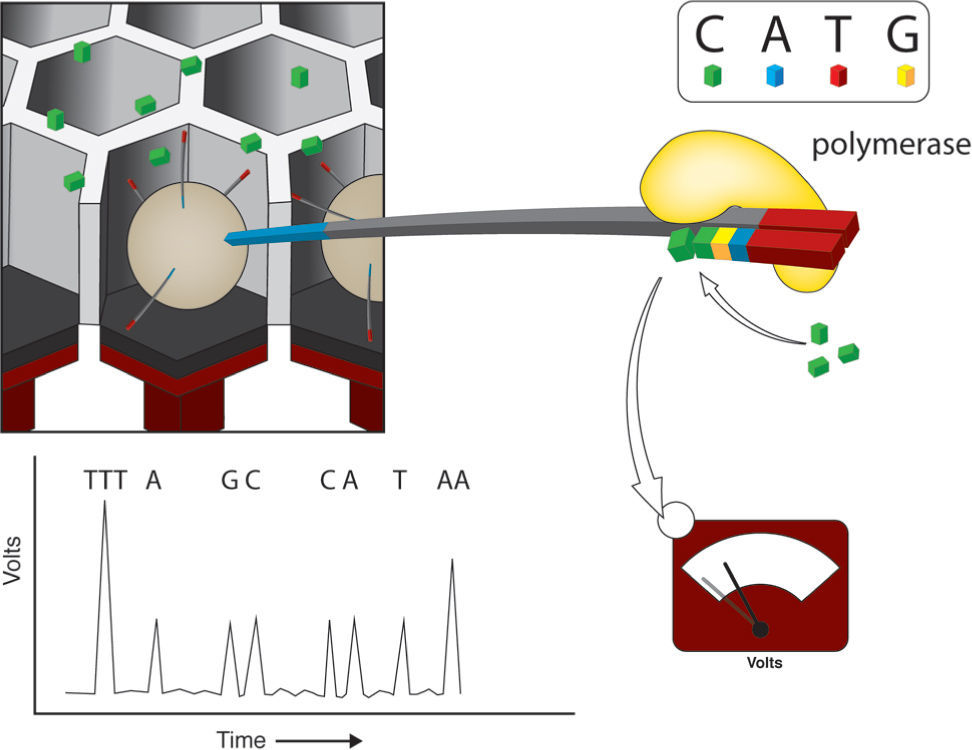
Ion-torrent (the chip is the machine). This platform is gaining popularity in the market mainly due to the low cost of sequencing runs and the possibility of in-house daily use without many technical requirements or maintenance. The library construction procedure is almost identical to 454 or pyrosequencing, including DNA fragmentation and adaptor ligation. The bead library from the emulsion PCR is deposited into a “chip” with millions of wells (165–600million depending on the specific version; Shokralla et al., 2012), where only 1 bead fits in each well. The extraordinary aspect of this technology is the chip itself. Each well is integrated to the chip's ion-sensitive layer and a proprietary ion sensor to register the very small voltage changes (per well) that result from nucleotide addition during DNA sequencing by synthesis (Rothberg et al., 2011, Fig. 4). As in 454, nucleotide addition is not followed by termination and it proceeds in cycles, 1 nucleotide after another. This results in the same problem that the pyrosequencing platforms have with homopolymer detection. The read length of ion Torrent is currently about 150bp with a final output of 3Gb per run (Liu et al., 2012), but it has increased up to 75 additional bp per read, and up to 10Gb per run (Shokralla et al., 2012; Table 1).
Ion-Torrent. The bead libraries derived from emulsion PCR are deposited into a “chip” with millions of cavities or wells that fit only one well. Each well is part of a microtransistor that integrates the chip, where voltage changes can be registered individually. Sequencing proceeds in a similar fashion as pyrosequencing, but instead of light emission, the addition of each base produces a voltage change. Modified form Niedringhaus et al. (2011).
Single molecule real time (SMRT) sequencing. Technological bets on massive sequencing are focused on the possibility to sequence single molecules in real time or “single molecule real time” (SMRT) sequencing. These technological developments are referred to as the third generation sequencing and, to date, only Pacific Biosciences (PacBio) has released a commercial platform implementing SMRT sequencing. SMRT implements the possibility of attaching a polymerase to a sequencing matrix and be able to follow in real time the synthesis process of a single DNA molecule (Fig. 5). An important feature of SMRT is that it does not require library construction as a prior step to sequencing, increasing the sequence production rate. Also, this technology allows for longer reads. The read length of Pacific Biosciences system has been reported to be up to 1 500bp with a final output of 6075Mb per run (Shokralla et al., 2012 ; Table 1). However, it is difficult to score single base additions in real time. The main problem in scoring real time single base additions is the high speed at which each polymerase synthesizes DNA exhibits stochastic fluctuations (Eid et al., 2009), thus, each enzyme has to be monitored individually and nucleotide additions registered at the appropriate speed in real time. This difficulties result in the highest error rates among NGS platforms (Table 1).
Single molecule real time sequencing (SMRT). PacBio. Single molecules of polymerase enzymes are attached to a sequencing matrix where just one DNA molecule will be synthesized. The enzymatic synthesis is followed individually, in real time, to reconstruct the sequence. SMRT technology does not require library construction as a prior step to sequencing, increasing the sequence production rate. Modified from Metzker (2010).
Currently, the main bottlenecks in genomic studies are in the post-sequence stages or assembly, which usually requires major computational capacities (Henson et al., 2012). Genomes sequences are variable (for instance, including heterozygosity in diploid organisms) and can be highly repetitive, and during assembly it is necessary to distinguish this “real” variation from sequencing errors and stay within reasonable computational times, making assembly a complex problem (Henson et al., 2012). The task would be much simpler if it could be determined whether a given set of reads corresponds to overlapping positions on the genome. In this sense, generally longer reads help to correctly find overlapping regions. Given the presence of short reads in all NGS platforms, it is clear that assembling genomes, metagenomes or transcriptomes is a task that faces enormous challenges, which are even more challenging due to high error rates.
The ease and accuracy of assembly depends on the degree of overlapping between reads. Two reads are considered as overlapping when there is a sequence match between reads that is long enough to be reliably distinguished from a random event (Henson et al., 2012). Thus, high uncertainty in assembly arises from locations in which not enough overlap is present to extend the genome sequence. In combination, coverage and read length increase confidence levels of the assembly process. Experience and models show that a good assembly using Sanger reads requires each base to be covered on average by at least 3 reads (3X, Lander and Waterman, 1988). However, for the short reads of NGS platforms, this number can rise up to 30X (Farrer et al., 2009; Meyer et al., 2012). High error rates also affect assembly and thus higher coverage is needed.
Published genomes have used between 5X and 10X with Sanger (Adams et al., 2000; Venter et al., 2001; Venter et al., 2004), but new publications have begun to report 50X, 100X and higher coverage with NGS (Diguistini et al., 2009; Quail et al., 2012). However, even with high coverage, overcoming the problem of repeats and derived assembly gaps sometimes need to be spanned by paired reads (2 reads generated from a single fragment of DNA and separated by known distance), which are available for most NGS platforms (Schatz et al., 2010). Much research has been published on the assembly problem, as well as development of new assembly algorithms and platforms (for recent reviews see Schatz et al., 2010; Nagarajan and Pop, 2013). Nevertheless, it has been suggested that the best approach is to use a reference genome sequence as a guide to resolve repeats, an approach known as “comparative assembly” (Pop et al., 2004).
A detailed analysis by Schatz et al. (2010) on the assembly of large genomes using NGS (Illumina and 454) showed that assemblies with these platforms are inferior than those accomplished using Sanger technology, but recognized the appeal of the lower costs of NGS. In this paper, guidelines are given to decide the best way to proceed in terms of choosing the platform and assembly method when pursuing a genome sequencing project. As we mention above, the keys to good assembly results from deep coverage by reads with lengths longer than common repeats, paired-end reads from short (0.5–3kb) and long (> 3kb) DNA fragments. Using NGS platforms, the most cost-effective way to obtain sequence coverage is to use pair-end sequencing by Illumina with at least 20X coverage. Schatz et al. (2010) also mention that with the assembly software available today, it is technically feasible and cost-effective to build a good assembly entirely from short reads (Illumina 300bp). A reliable genome project should be planned with the aim of producing deep coverage (30X) in paired-end sequences from short DNA fragments (0.5–1kb) and additional coverage (10–20X) in paired ends from longer DNA fragments (3–10kb). A similar strategy was followed to produce the panda genome (Ruiqiang Li et al., 2010). Another robust option would be to combine platforms, as implemented in the turkey genome assembly (Dalloul et al., 2010), in which lower coverage (5X with 454) and higher coverage (25X with Illumina) approaches are simultaneously used to overcome the coverage-ease of assembly paradox.
Considerations on information managementThe size of data outputs from NGS platforms represent an important challenge in terms of information processing and analysis (Schuster, 2008; Liu et al., 2012; Yoccoz, 2012), which has triggered advancement in information and data management technologies. In a recent opinion published by Yoccoz (2012), a critical view is presented on the analytical challenges posed by massive data in the study of microorganism biodiversity (inventories) from environmental samples. This view is also applicable to other types of studies, such as whole genome and transcriptome sequencing. The author identifies 2 major obstacles when dealing with such data: i) data storage and management, and ii) implementation and assessment of statistical models.
Regarding data storage and management, the challenges are very clear: the amount of data that are produced by each platform per run ranges from 1 to 600GB (Table 1). Thus, the processing, management and storage of data from multiple projects becomes a major task and needs the development of a sophisticated system to handle information, from sample labeling to library construction (multiplexing labeling), as well as a system of sequencing and informatic analysis (Liu et al., 2012). Most laboratories will outsource library construction and sequencing. However, sample and data management, as well as bioinformatic processing and analyses will usually be in-house tasks. The size of data sets that are generated in NGS is also an issue for sorting specific information and for the implementation and assessment of statistical models. Thus, following Cardenas and Tiedje (2008) we emphasize that “the limitation is not the ability to produce sequence data but the ability to store and analyze it in new revealing ways”.
Platforms: which one is best?All sequencing platforms have been advancing towards massive DNA sequencing of longer DNA fragments, and to the production of even larger data sets. To take the full advantage of the generated data, equally massive computational tools are required. However, these computational requirements vary from platform to platform, thus it is important to evaluate the available bioinformatic resources (e.g., hardware, software, human resources) before deciding to choose one technology over another. The bioinformatics challenge is of such relevance for the development of current and future sequencing technologies, that it has triggered a revolution in the information technologies and massive data storage (Mardis, 2008; Henson et al., 2012).
Error rates. All massive sequencing platforms have relatively high error rates (compared with traditional Sanger technology). In addition, each platform has specific types of error associated with the sequencing protocol. The type and rate of error have consequences in the informatic processing of the data, as they need to be identified and “cleaned” before data analyses per se (Mardis, 2008). Even after being “cleaned”, these errors can have consequences during the assembly of fragments. As a result of the different types of error between platforms, the common suggestion is to simultaneously use 2 different platforms for the assembly of complete genomes (Aury et al., 2008; Dalloul et al., 2010). Also, in cases where the error rates are high or high diversity of sequences is expected, it is recommended to compensate with a high coverage per sequencing sample.
When to pick which? As we have reviewed in this paper, coverage, length of reads, error rates, and costs will vary depending on the sequencing platform, making some of these more suitable than others for specific studies (Table 1). In the absence of reference genomes or in non-model organisms, genome sequencing (de novo sequencing) and assembly, as well as characterization of transcriptomes, can be a daunting task from a bioinformatic point of view. Thus, the assembly process benefits greatly from longer reads and higher coverage, both offered by platforms such as pyrosequencing and synthesis, respectively (Glenn, 2011; Martin and Wang, 2011; Cahais et al., 2012). When reference genomes are available, a good alternative when performing experiments and evaluating transcriptome results would be those technologies that, even with short reads, have a sufficiently high coverage to use a reference genome as a scaffold.
In terms of number of publications, sequencing by synthesis using Illumina is dominant (http://www.illumina.com/science/publications-list.ilmn). This platform has been used in studies focused on characterizing transcriptomes, re-sequencing entire genomes and in studies that look for single nucleotides polymorphisms (SNPs), because it is possible to verify that the mutations found are not a product of sequencing errors (Li et al., 2013). The steady increment in read length for Illumina platforms (Shokralla et al., 2012) has made it appropriate for de novo assembly of genomes (Dalloul et al., 2010; Li et al., 2010) or transcriptomes (Martin and Wang, 2011), particularly if used in parallel with other platforms that provide longer reads. Moreover, given the development of new software tools and pipelines (e.g. SHARE, Rodrigue et al., 2010), Illumina technologies are also being used in metagenomic analyses (Coetzee et al., 2010; Rodrigue et al., 2010).
Choosing sequencing approaches for biodiversity inventories or monitoring should be done considering the number of individuals and the portion of the genome to be sequenced. If macroscopic organisms are to be identified via barcoding sequences, it is usually more practical to follow a Sanger approach because specific sequences for only a few individuals are needed. In contrast, inventories for microorganisms have been performed following a metagenomic approach, where entire communities are sequenced for a conserved region that can be used to identify taxonomic groups. To date, pyrosequencing have been traditionally used in microbial ecology studies (Sogin et al., 2006). Given the vast diversity usually found in microbial communities, approaches that give more coverage are being used more recently (Degnan and Ochman, 2012). Nonetheless, given the number of publications, we observe that in metagenomic studies, pyrosequencing is the preferred choice above other approaches with shorter reads (http://454.com/publications/all-publications.asp). However, Roche recently announced the shutdown of pyrosequencing platforms (http://www.genomeweb.com/sequencing/roche-shutting-down-454-sequencing-business), which would probably trigger the use of Illumina or platforms such as Ion Torrent.
Although platforms such as Ion Torrent and SOLiD are in the market, publications using these approaches are notably scant in comparison to 454 or Illumina. In the case of Ion Torrent, this could partly be the result of their recent availability in the market. Ion Torrent equipment is being sold massively and it is possible that many publications will appear soon, probably in the field of metagenomics. In the case of SOLiD, its limited impact (in terms of the number of publications) may be the result of the complexity of data processing and assembly (Flicek and Birney, 2010). Nonetheless, SOLiD is recognized as a very reliable platform in the characterization of SNPs and transcriptomes, and in the quantification of mRNA (Glenn, 2011). Furthermore, this platform has been recently used for de novo sequencing of the pig genome (Rubin et al., 2012).
Besides genome sequencing, other options exists where NGS can be used to study genetic variation of entire populations of non-model organisms, for which funding may be limited or research questions are not directed towards full genome sequencing. There are some strategies that take advantage of NGS for SNP detection without the need for assembly and that can be used to simultaneously screen genetic variation of entire populations. One of these strategies is known as RAD-tag sequencing, and it seems to be a productive and promising avenue in population genetics (Baird et al., 2008). Restriction-site Associated DNA (RAD) markers are short fragments of DNA adjacent to a particular restriction enzyme recognition site. This approach can be linked to the use of microarrays and hybridization to screen thousands of polymorphic markers. The use of NGS platforms, such as Illumina, now allows to screen, sequence and detect particular SNPs within the RAD-tag fragments generated, facilitating the rapid discovery of thousand SNPs and high throughput of many populations (Baird et al., 2008). Thus, RAD-seq combines molecular biology techniques with Illumina sequencing: enzymatic restriction of DNA (as for RFLPs and AFLPs) and the use of molecular identifiers (MID) to associate sequence reads to particular individuals (Davey et al., 2010). RAD-tag sequencing has been found to be particularly useful in studies of wild populations and non-model study species, promising to become a practical approach taken in ecological population genomics (Davey et al., 2010). The main reasons for the popularity of RAD-tag sequencing and similar methods are that: i) there is no variant discovery assay step; ii) it is a very versatile method, expected to work with any restriction enzyme or any species; iii) it can be applied to many research problems, from identifying SNPs at large-scale population genotyping to create linkage maps of Mendelian or quantitalive trait loci (QTL).
NGS: opportunities and challenges for the study of biodiversityThe advent of high throughput sequencing technologies has made genomics not the end itself, but an ideal tool to advance in the study of biodiversity and related processes with relative ease and much greater detail and depth than ever before. The questions that can be addressed using NGS technologies range from classical population genetics to community ecology, passing through phylogeography (McCormack et al., 2013), historical demography (Pool et al., 2010; Li and Durbin, 2011), molecular systematics and phylogenetics (Parks et al., 2009). In particular, the NGS methods allow a more detailed and comprehensive description of biodiversity at any level of biological organization than any previous technology (Taberlet et al., 2012), information that can be used to design conservation and management strategies with a more systemic vision.
Studies on population genetics and molecular evolution have historically been limited by the amount of genetic variation that can be accessed with traditional gene-by-gene approaches (Eguiarte et al., 2013). In addition, the fragmented genomic information coming out from these approaches limits our capacity to determine key parameters, such as recombination, substitution rates and demographic changes (Hedrick, 2000). The acquisition of tens to hundreds of markers and genome data for non- model organisms through classical or Sanger sequencing becomes both costly and slow, while high throughput sequencing makes possible (budget and technically wise) to scan entire genomes of non-model organisms to perform comparative and population genomics studies (Hohenlohe et al., 2010). Such studies have been mainly applied to domesticated species of plants and animals (Groenen et al., 2012; Qin et al., 2014). Some studies, however, have dealt with wild organisms, resulting in the identification of the genetic basis of adaptation (for example see studies on the 3-spine stickle back fish, (Hohenlohe et al., 2010); a tropical lizard, (Freedman et al., 2010); and the wild maize (teosinte), (Eguiarte et al., 2013; Pyhäjärvi et al., 2013)).
A common challenge for these type of studies involves the mere size of the genomes of many organisms, specially those with relatively big, polyploid genomes (Niklas, 1997; Amborella Genome Project, 2013). Additionally, the genomes of many organisms are very dynamic, due to the high levels of recombination, the presence of repetitive non-coding regions and mobile elements and there can be wide variance in the genome sizes of a given species (Chia et al., 2012; Díez et al., 2013). In many cases, these difficulties can be tackled with a low coverage transcriptome or by reducing the number of studied genomic regions (e.g., RAD-tag, see above). These 2 approaches are good starting options to search for genome-wide genetic variation in non-model organisms, and to estimate population genetics parameters, such as genetic structure, effective population sizes, gene flow, inbreeding depression and natural selection (Allendorf et al., 2010; Hohenlohe et al., 2010; Andrew et al., 2012; Ashrafi et al., 2012; Hill et al., 2013). Furthermore, transcriptome data can be used in functional studies to compare patterns of gene expression related to environmental conditions, sexes, life history stages and different structures of an organism (Ekblom and Galindo, 2011). The same genomic and transcriptomic approaches can be employed to identify regions suitable to study phylogenetic relationships, particularly between closely related species or intra-specific varieties, for instance the study of Eaton and Ree (Eaton and Ree, 2013) using RAD-seq (a variant of RAD-tag method described above) data in a small group of Pedicularis (Orobanchaceae) plant species from Tibet. Whole plastome sequencing in pines has been used to evaluate divergence among closely related species (Parks et al., 2009) and between small endemic populations of North American pines (Whitall et al., 2010).
NGS technologies can also be used to describe microbial species or groups of species present in complex biological samples [e.g., tissue (Kemler et al., 2013), soil (López-Lozano et al., 2013), or water samples (Zaremba-Niedzwiedzka et al., 2013)]. These studies are generally known as metagenomic, and they can look at specific genomic signatures or entire genomes of communities without the need to culture or isolate microbial species in the laboratory. The culture independent metagenomic approach using NGS opens a wide new window for microbiology, allowing us to go way beyond the less than 5% of the estimated microbial diversity that can be successfully characterized using traditional culturing methods (Torsvik et al., 1990). A simple metagenome would involve the NGS of 16S ribosomal RNA, or other highly conserved gene, that can serve to build a coarse inventory of diversity of a given sample (Sogin et al., 2006). Given the sequencing capacity that NGS platforms offer, meta-genomes can be easily extended to look for multiple genes or entire genomes (Bonilla-Rosso et al., 2008; for recent studies in Cuatro Ciénegas in Mexico see Bonilla-Rosso et al., 2012 and Peimbert et al., 2012). Experimental designs looking at different samples and treatments, either across space or time, promise to “unlock the potential of metagenomics” (Knight et al., 2012), which can greatly advance our understanding of the processes occurring at the microbial level (Kahvejian et al., 2008). These NGS technologies have already revolutionized our understanding of the microbial diversity of the planet (Sogin et al., 2006; Lauber et al., 2009; Martiny et al., 2011) and even that occurring within our bodies (Costello et al., 2012; Human Microbiome Consortium, 2012). Nonetheless, challenges remain, such as taxon-biased DNA extraction, intrinsic sequencing errors, PCR primers biases (in the case of 16S rDNA approaches), overestimation of taxon abundance or distinguishing meta-genomic signatures of uncultured taxa from artifacts (chimeras).
ConclusionsThe main revolution and challenge with the availability of massively parallel sequencing technologies is the production of enormous amounts of data in relatively short times. The methodological differences for each platform bring along different advantages and limitations that impact their use in different studies. Thus, a key step is to identify the features that each platform has in order to make a better choice when designing research projects. Finally, given the amount of information generated by these technologies we must stress the importance of the need for computational resources and efficient pipelines for data processing, because these can be the major limitation in the success of a research project based on massive sequencing technologies.
This review is a result from the workshop “Avances y perspectivas del uso de métodos de secuenciación de próxima generación en el studio de la genética de poblaciones y su empleo en problemas ambientales”, held in México City at Instituto de Ecología in January 2013, and funded by Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. AEE wants to acknowledge financial support from AMC-Loreal-UNESCO scholarship (2012), UC-Mexus collaborative grant (2013) and PAPIIT-UNAM IA200814 (2014). LEE also wants to thank the support of the grants Conacyt CB2011/167826, Sep-Conacyt-ANUIES-ECOS Francia M12-A03, PAPIIT UNAM IN202712 and Conabio KE 004 for NGS related recent research.