All the currently recognized Malassezia species have been isolated from mammals. However, only a few of them have been isolated from birds. In fact, birds have been less frequently studied as carriers of Malassezia yeasts than mammals.
AimIn this study we describe two new taxa, Malassezia brasiliensis sp. nov. and Malassezia psittaci sp. nov.
MethodsThe isolates studied in this publication were isolated from pet parrots from Brazil. They were characterized using the current morphological and physiological identification scheme. DNA sequencing and analysis of the D1/D2 regions of the 26S rRNA gene, the ITS-5.8S rRNA gene sequences and the β-tubulin gene were also performed.
ResultsThe strains proposed as new species did not completely fit the phenotypic profiles of any the described species. The validation of these new species was supported by analysis of the genes studied. The multilocus sequence analysis of the three loci provides robust support to delineate these species.
ConclusionsThese studies confirm the separation of these two new species from the other species of the genus Malassezia, as well as the presence of lipid-dependent Malassezia yeasts on parrots.
Todas las especies del género Malassezia actualmente identificadas se han aislado de mamíferos. Sin embargo, tan solo unas pocas de ellas se han aislado de aves. De hecho, las aves han sido estudiadas con menos frecuencia como portadoras de estas levaduras que los mamíferos.
ObjetivosEn este estudio describimos dos nuevas especies del género Malassezia: Malassezia brasiliensis sp. nov. y Malassezia psittaci sp. nov.
MétodosLas cepas estudiadas en esta publicación se aislaron de loros utilizados como animales de compañía en Brasil. Las cepas se caracterizaron mediante los criterios morfológicos y fisiológicos actualmente utilizados para la identificación de estas levaduras. También se llevó a cabo la secuenciación y el análisis de los fragmentos génicos D1/D2 26S e ITS-5.8S del ADN ribosómico y del gen de la β-tubulina.
ResultadosLos perfiles fenotípicos de las cepas propuestas como nuevas especies no encajaron completamente con los de las especies descritas en este género. Además, el análisis de los genes estudiados respaldó la validez de las nuevas especies. El análisis multilocus de secuencias de los tres loci estudiados reforzó con mayor firmeza la definición de las nuevas especies.
ConclusionesTodos estos estudios confirman la separación de estas dos nuevas especies del resto de las especies descritas del género Malassezia, así como la existencia de especies dependientes de lípidos del género Malassezia en loros.
Malassezia species are lipophilic basidiomycetous yeasts that inhabit the skin and mucosal sites of a variety of homoeothermic animals, such as mammals or birds. In some conditions, these yeasts can be opportunistic pathogens of humans and animals.29 This monophyletic genus is the only genus included in the order Malasseziales, which has an uncertain taxonomic position in the subphylum Ustilagomycotina (e.g., smut fungi). Recently, the class Malasseziomycetes has been proposed to accommodate these fungi.31 On the other hand, some culture-independent studies of fungi from environmental samples (e.g. Antarctic soils, corals, deep-sea sediments, nematodes) have shown that Malassezia are exceedingly widespread and ecologically diverse.1 However, these yeasts have been only recovered from mammals and birds at the moment.
Nowadays Malassezia yeasts have been isolated mainly from domestic animals, different wild animals held in captivity, and also from wildlife.29 However, the occurrence of Malassezia yeasts on the skin of most animals remains unknown. At present, the genus Malassezia includes 14 species. Most of them are lipid-dependent yeasts which require long-chain fatty acids to grow, while the lipophilic but non-lipid-dependent species Malassezia pachydermatis is the only species in the genus that does not require lipid supplementation for development in culture medium.5
These species have been isolated mainly from mammals, and only a few of them have been isolated from birds. In fact, birds have been less frequently studied as carriers of Malassezia yeasts than mammals. Therefore, few papers have been published about these yeasts from birds. A search in PubMed (www.ncbi.nlm.nih.gov/pubmed; US National Library of Medicine) early in 2015 yielded more than 1500 journal citations containing the keywords “Malassezia AND mammals”, but only 7 using the keywords “Malassezia AND birds”.
Besides, little is known about Malassezia yeasts isolated from birds.29 At the moment, only M. pachydermatis, Malassezia furfur and Malassezia sympodialis have been reported from these animals. As far as M. pachydermatis is concerned, this non lipid-dependent species has been reported to be isolated from feathers of a variety of birds,11 from a diseased throat of a scarlet macaw2 and from several wild bird droppings.23 Some strains of the lipid-dependent species M. furfur were isolated from the wing and mouth of ostriches and from the wing of a pelican,15 and also from pigeon droppings.8 The lipid-dependent species M. sympodialis was the most frequently yeast isolated from clinically altered combs of adult chickens.18 In this study, the isolates were identified only in the basis of phenotypic characteristics without DNA sequencing confirmation. However, Malassezia yeasts have been isolated from birds, including parrots, in a previous study.24 Nonetheless, in this study, the isolates were not identified at species level. On the other hand, some studies have failed to demonstrate the presence of lipophilic yeasts in these animals. Malassezia yeasts were not detected on the skin of healthy psittacine birds and neither on psittacine birds with feather-destructive behavior.25
In this paper, we describe two new lipid-dependent species in the genus Malassezia isolated from parrots. For these isolates we propose the names Malassezia brasiliensis sp. nov. and Malassezia psittaci sp. nov.
Materials and methodsStrainsThe strains proposed as new species in this publication were isolated from pet parrots from Brazil. Four isolates were recovered in the Laboratory of Molecular and Cellular Biology of the Paulista University (São Paulo, Brazil) from four individual parrots (Table 1). They were isolated from lesions on beak (MA 1453, Amazona aestiva; MA 1454, Pionus menstruus; MA 1455, A. aestiva) and oropharynx (MA 1456, Nymphicus hollandicus) in these animals. Unfortunately the etiological significance of these isolates in these lesions was not determined. The strains were stored at −80°C.9
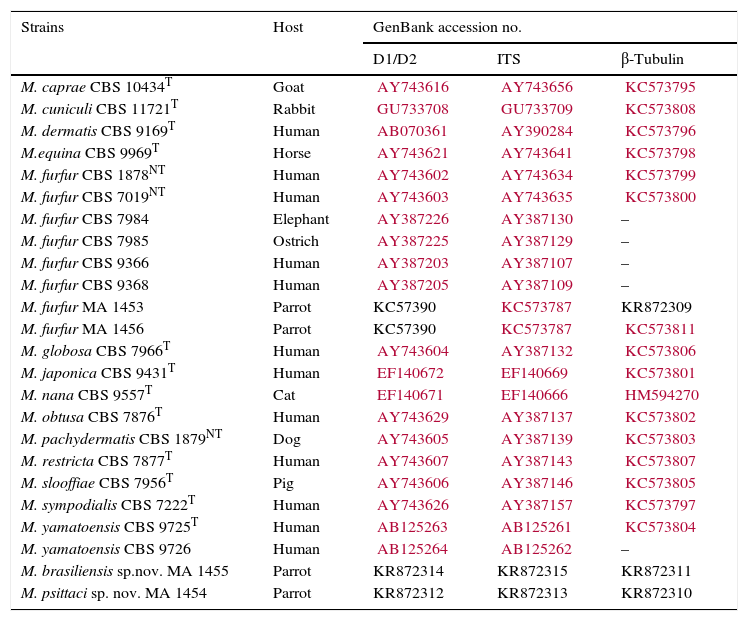
Strains studied, their hosts and GenBank accession numbers.
| Strains | Host | GenBank accession no. | ||
|---|---|---|---|---|
| D1/D2 | ITS | β-Tubulin | ||
| M. caprae CBS 10434T | Goat | AY743616 | AY743656 | KC573795 |
| M. cuniculi CBS 11721T | Rabbit | GU733708 | GU733709 | KC573808 |
| M. dermatis CBS 9169T | Human | AB070361 | AY390284 | KC573796 |
| M.equina CBS 9969T | Horse | AY743621 | AY743641 | KC573798 |
| M. furfur CBS 1878NT | Human | AY743602 | AY743634 | KC573799 |
| M. furfur CBS 7019NT | Human | AY743603 | AY743635 | KC573800 |
| M. furfur CBS 7984 | Elephant | AY387226 | AY387130 | – |
| M. furfur CBS 7985 | Ostrich | AY387225 | AY387129 | – |
| M. furfur CBS 9366 | Human | AY387203 | AY387107 | – |
| M. furfur CBS 9368 | Human | AY387205 | AY387109 | – |
| M. furfur MA 1453 | Parrot | KC57390 | KC573787 | KR872309 |
| M. furfur MA 1456 | Parrot | KC57390 | KC573787 | KC573811 |
| M. globosa CBS 7966T | Human | AY743604 | AY387132 | KC573806 |
| M. japonica CBS 9431T | Human | EF140672 | EF140669 | KC573801 |
| M. nana CBS 9557T | Cat | EF140671 | EF140666 | HM594270 |
| M. obtusa CBS 7876T | Human | AY743629 | AY387137 | KC573802 |
| M. pachydermatis CBS 1879NT | Dog | AY743605 | AY387139 | KC573803 |
| M. restricta CBS 7877T | Human | AY743607 | AY387143 | KC573807 |
| M. slooffiae CBS 7956T | Pig | AY743606 | AY387146 | KC573805 |
| M. sympodialis CBS 7222T | Human | AY743626 | AY387157 | KC573797 |
| M. yamatoensis CBS 9725T | Human | AB125263 | AB125261 | KC573804 |
| M. yamatoensis CBS 9726 | Human | AB125264 | AB125262 | – |
| M. brasiliensis sp.nov. MA 1455 | Parrot | KR872314 | KR872315 | KR872311 |
| M. psittaci sp. nov. MA 1454 | Parrot | KR872312 | KR872313 | KR872310 |
CBS, Centraalbureau voor Schimmelcultures; MA, culture collection of the Veterinary Mycology group.
Prefixes KR of accession numbers correspond to the sequences generated in this study.
The characterization of lipid-dependent yeasts was based on their inability to grow on Sabouraud's glucose agar (SGA; Oxoid, Basingstoke, UK), on their ability to use certain polyoxyethylene sorbitanesters (Tweens 20, 40, 60 and 80), and the use of additional tests such as the Cremophor EL assimilation test, or the splitting of esculin (β-glucosidase activity), following the identification schemes of Guého et al.14 Other tests, such as the catalase reaction, growth at different temperatures (32°C, 37°C, 40°C and 45°C) on modified Dixon agar (mDA; 36g malt extract, 10g peptone, 20g desiccated ox-bile, 10ml Tween 40, 2ml glycerol, 2ml oleic acid and 12g agar per liter, pH 6.0), and the morphological characteristics after incubation at 32°C for 7 days in the same culture medium were also performed.14
DNA extraction, gene amplification, sequencing and phylogenetic analysisDNA of the isolates MA 1453, MA 1454 and MA 1455 was extracted and purified directly from seven day-old cultures on modified Dixon agar according to the FastDNA Spin kit protocol with the FastPrep FP-24 instrument (MP Biomedicals, Biolink, Barcelona, Spain). The DNA was kept at −20°C until used as a template for PCR amplification. Sequences of D1/D2 26S rRNA (D1/D2), ITS-5.8S rRNA (ITS) and β-tubulin (β-tub) genes of currently recognized species and the isolate MA 1456 were obtained from previous studies.3,4,6 D1/D2, ITS, and β-tub genes of the strains isolated from parrots were amplified and sequenced as described previously.6
Sequence alignments were performed using the software program Clustal X v2.0.12.22 Regions of ambiguous alignment were removed with Gblocks.7 Parsimony analyses of the individual and combined data matrices were conducted using PAUP* version 4.0b10 software.30 One hundred heuristic searches were conducted with random sequence addition and tree bisection reconnection branch-swapping algorithms, collapsing zero-length branches and saving all minimal-length trees (MulTrees) on different data sets. The gaps were treated as missing data, and support of internal branches was assessed using heuristic parsimony search of 1000 bootstrapped data sets. Tree length, consistency index (CI), retention index (RI) and the homoplasy index (HI) values were also calculated. The combined data set was tested for incongruence with the partition homogeneity test (PHT) as implemented in PAUP*.
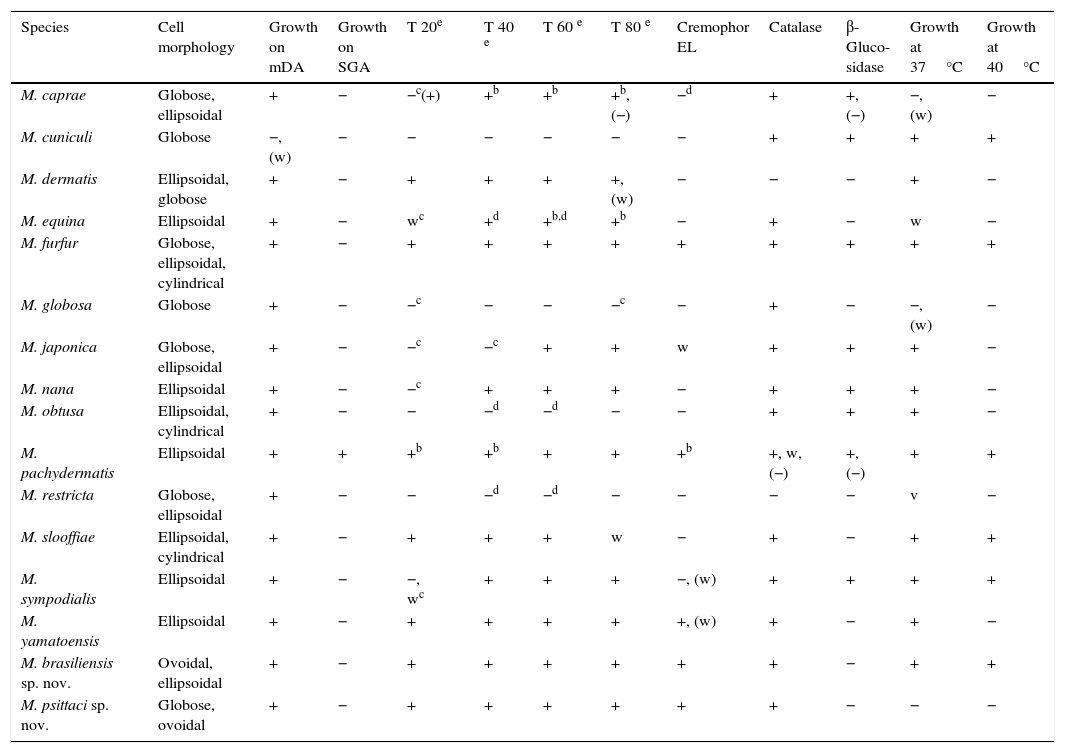
ResultsMorphology and physiologyThe phenotypic characteristics of the new species, M. brasiliensis (MA 1455) and M. psittaci (MA 1454) and the other currently accepted Malassezia species, are summarized in Table 2. None of these isolates grew on SGA without any lipid supplementation. M. brasiliensis cells were ovoidal to ellipsoidal (Table 2), and M. psittaci cells were globose to ovoidal (Table 2).
Main phenotypical characteristics of Malassezia species.a
| Species | Cell morphology | Growth on mDA | Growth on SGA | T 20e | T 40 e | T 60 e | T 80 e | Cremophor EL | Catalase | β-Gluco-sidase | Growth at 37°C | Growth at 40°C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. caprae | Globose, ellipsoidal | + | − | −c(+) | +b | +b | +b, (−) | −d | + | +, (−) | −, (w) | − |
| M. cuniculi | Globose | −, (w) | − | − | − | − | − | − | + | + | + | + |
| M. dermatis | Ellipsoidal, globose | + | − | + | + | + | +, (w) | − | − | − | + | − |
| M. equina | Ellipsoidal | + | − | wc | +d | +b,d | +b | − | + | − | w | − |
| M. furfur | Globose, ellipsoidal, cylindrical | + | − | + | + | + | + | + | + | + | + | + |
| M. globosa | Globose | + | − | −c | − | − | −c | − | + | − | −, (w) | − |
| M. japonica | Globose, ellipsoidal | + | − | −c | −c | + | + | w | + | + | + | − |
| M. nana | Ellipsoidal | + | − | −c | + | + | + | − | + | + | + | − |
| M. obtusa | Ellipsoidal, cylindrical | + | − | − | −d | −d | − | − | + | + | + | − |
| M. pachydermatis | Ellipsoidal | + | + | +b | +b | + | + | +b | +, w, (−) | +, (−) | + | + |
| M. restricta | Globose, ellipsoidal | + | − | − | −d | −d | − | − | − | − | v | − |
| M. slooffiae | Ellipsoidal, cylindrical | + | − | + | + | + | w | − | + | − | + | + |
| M. sympodialis | Ellipsoidal | + | − | −, wc | + | + | + | −, (w) | + | + | + | + |
| M. yamatoensis | Ellipsoidal | + | − | + | + | + | + | +, (w) | + | − | + | − |
| M. brasiliensis sp. nov. | Ovoidal, ellipsoidal | + | − | + | + | + | + | + | + | − | + | + |
| M. psittaci sp. nov. | Globose, ovoidal | + | − | + | + | + | + | + | + | − | − | − |
All the isolates studied have good growth on mDA at 32°C. The isolates MA 1453 (2.4–5.1mm in diameter; average diameter=4.2mm) and MA 1456 (3.6–6.4mm in diameter; average diameter=5.4mm) grew faster and formed larger colonies than MA 1455 (1.6–3.2mm in diameter; average diameter=2.5mm) and MA 1454 (1.9–3mm in diameter; average diameter=2.6mm) on mDA at 32°C after 7 days of incubation. With the exception of MA 1454 (M. psittaci) that did not grow at 37°C, the rest of the isolates showed good growth at both 37°C and 40°C after 7 days of incubation, but did not grow at 45°C. All the isolates had similar good growth around Tweens 20, 40, 60 and 80, and Cremophor EL. All, except one (MA 1453), were β-glucosidase negative.
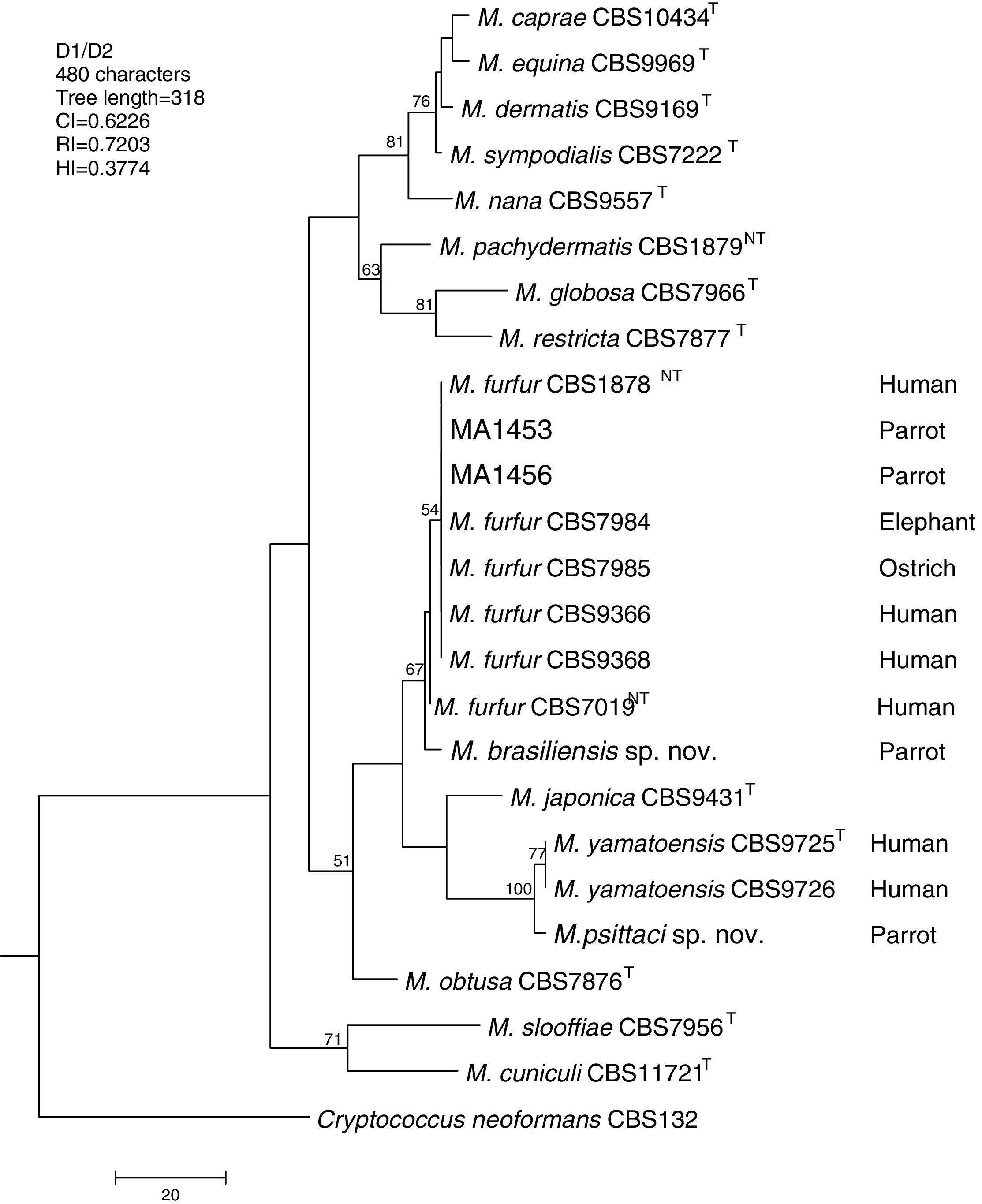
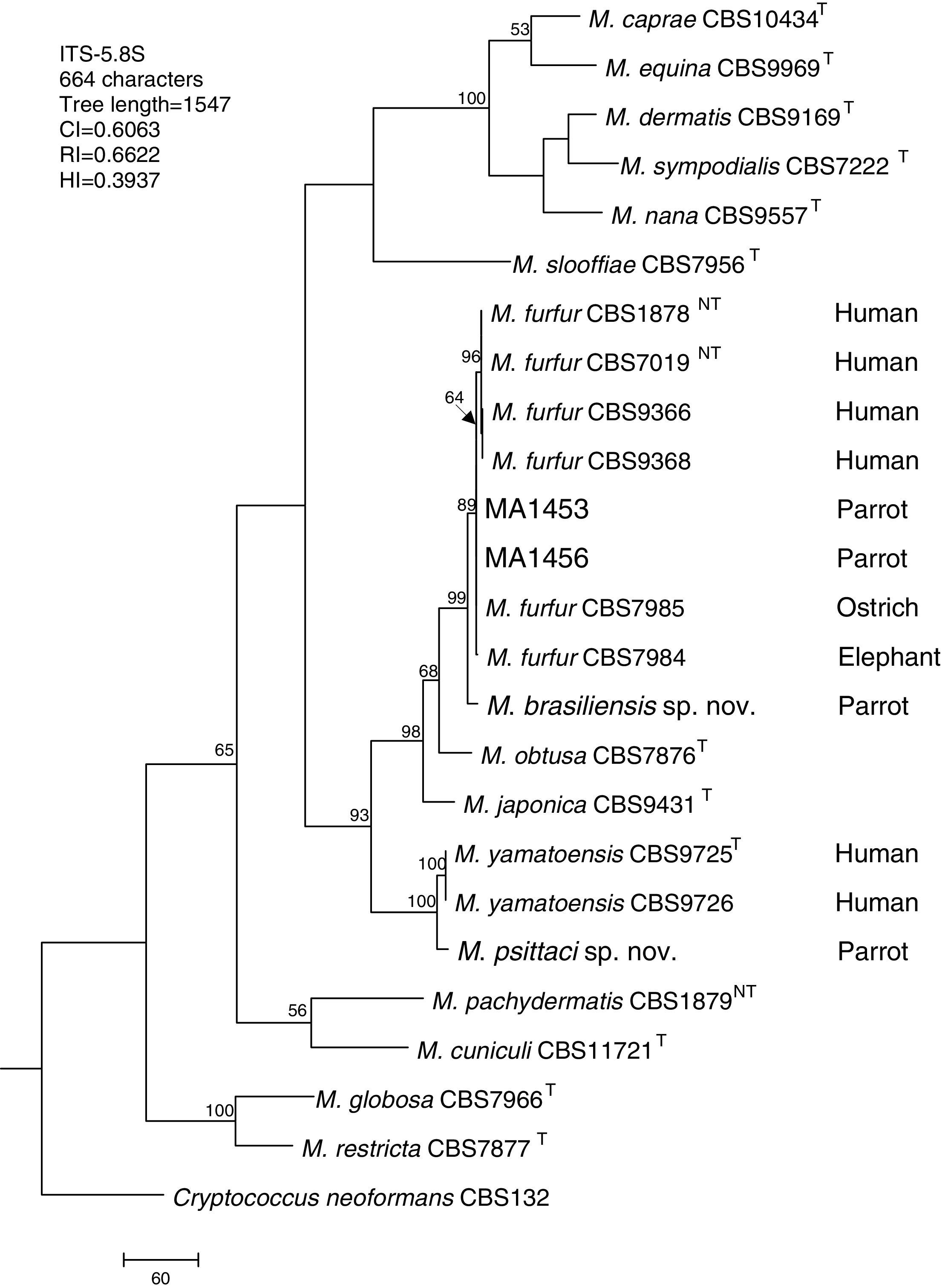
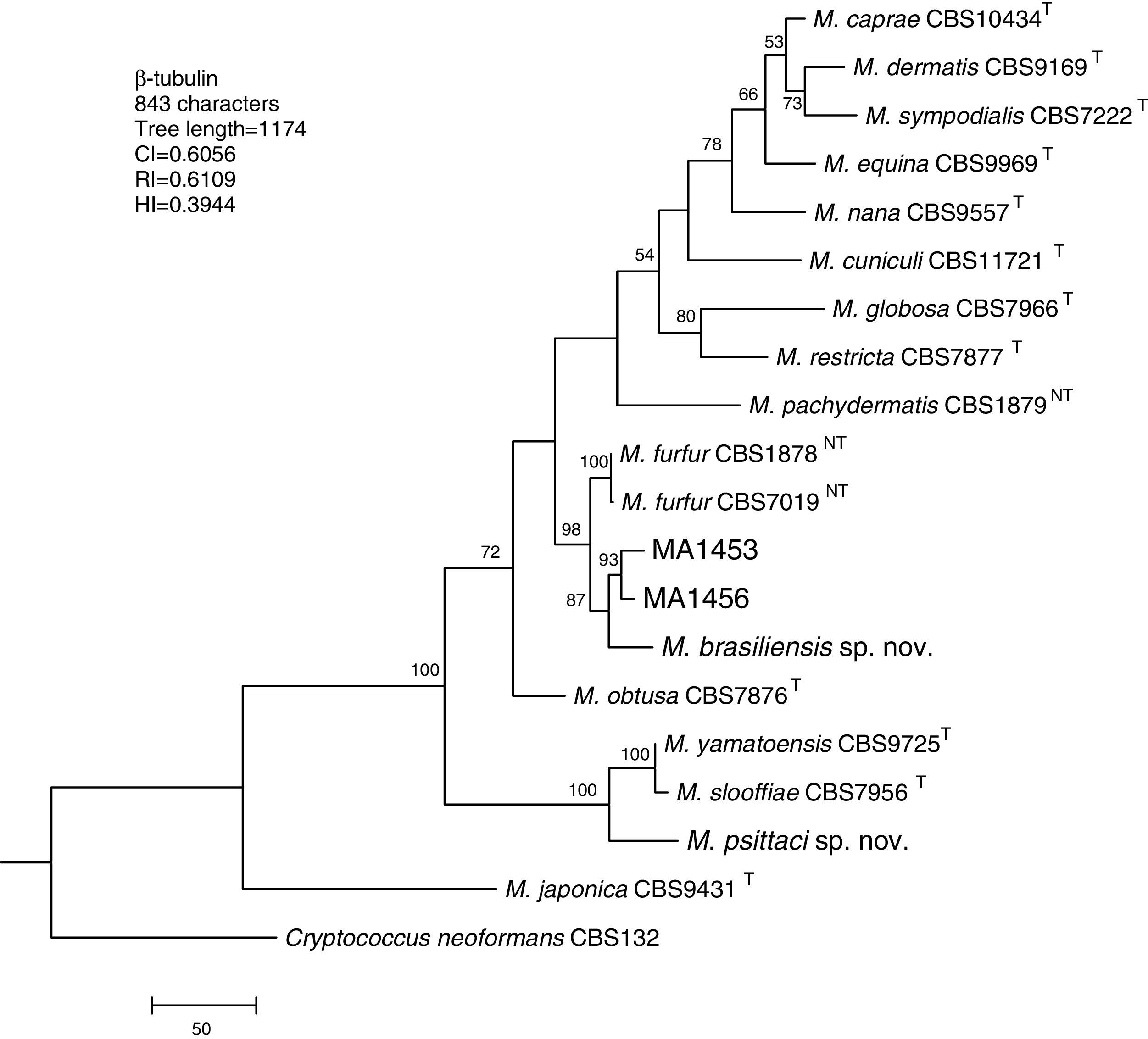
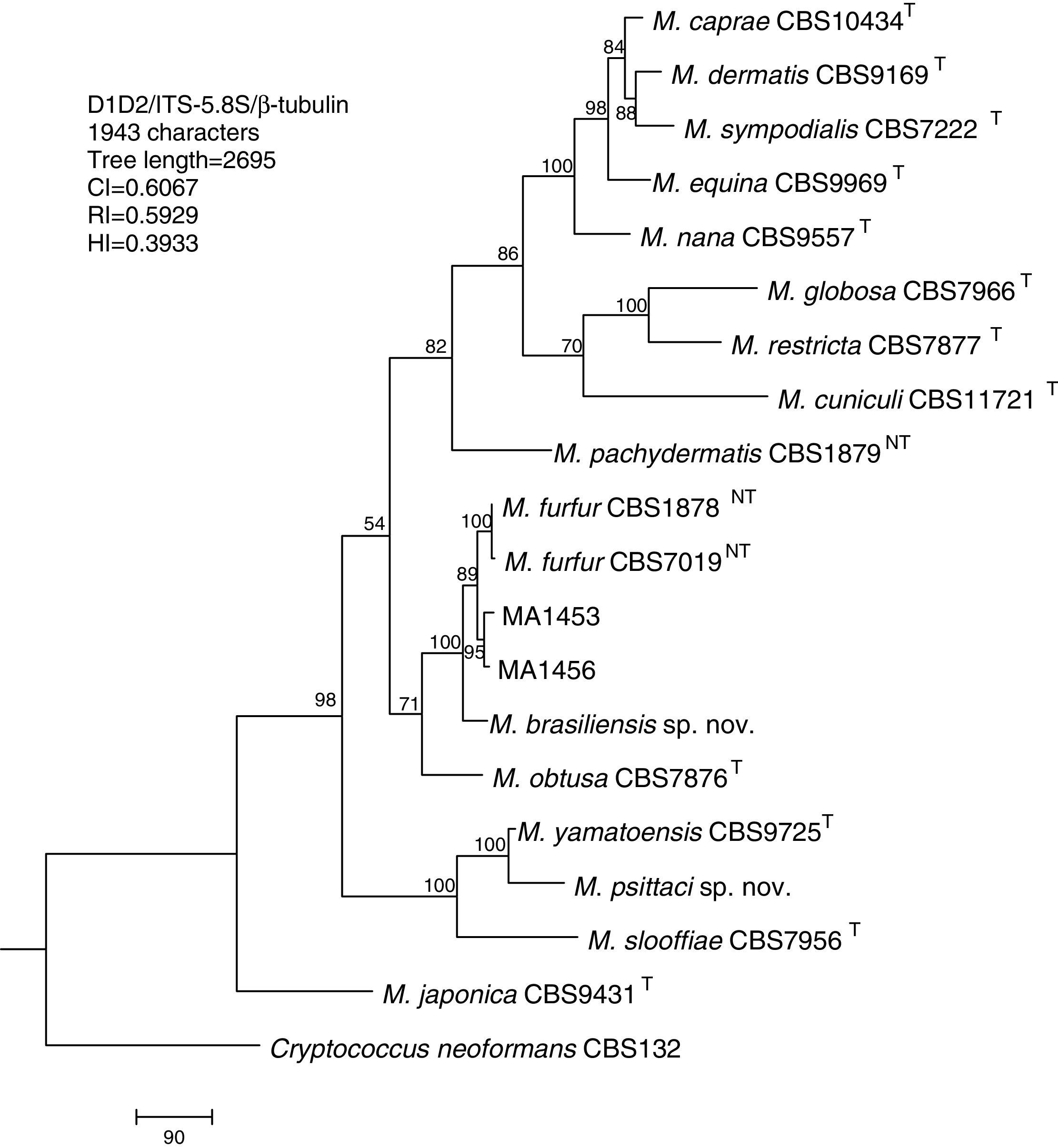
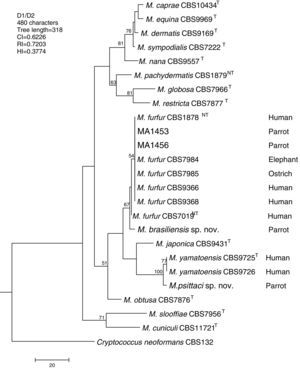
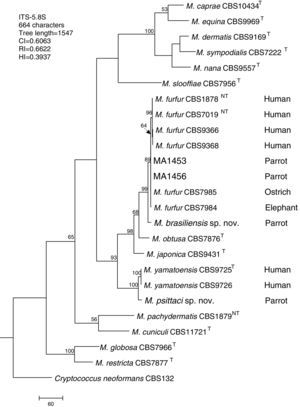
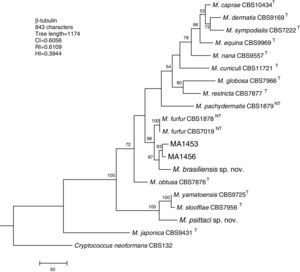
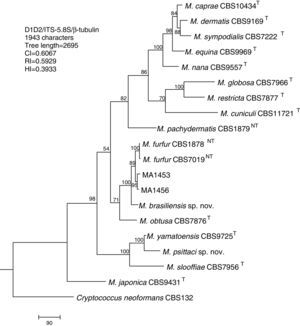
Molecular and phylogenetic analysisWith the primers used we were able to amplify and sequence 619bp, 717–829bp and 950–1115bp of the D1/D2, the ITS and the β-tub genes, respectively. The nucleotide sequences determined in this study have been deposited at the GenBank database under accession numbers KR872309–KR872315 (Table 1). Figs. 1, 2 and 3 show the molecular phylogenetic trees based on the maximum parsimony analysis of the sequences of D1/D2, ITS and β-tub, respectively. When the three loci were combined (Fig. 4), the data set included 1943 characters. Each sequence contributed to that length as follows: D1/D2, 501 characters; ITS, 599 characters; and β-tub, 843 characters. From these characters, 921 were constant, and 643 were parsimony informative (D1/D2 84, ITS 263 and β-tub 296). The result of the partition homogeneity test (p=0.01) showed that the data set could be combined without reducing phylogenetic accuracy.10
Molecular phylogenetic tree inferred from a parsimony analysis of D1/D2 26S rRNA sequences of members of the genus Malassezia. MP bootstrap values >50% in 1000 replications are shown at nodes. The tree is rooted with Cryptococcus neoformans. CI, consistency index; RI, retention index; HI, homoplasy index.
Molecular phylogenetic tree inferred from a parsimony analysis of ITS-5.8S rRNA sequences of members of the genus Malassezia. MP bootstrap values >50% in 1000 replications are shown at nodes. The tree is rooted with Cryptococcus neoformans. CI, consistency index; RI, retention index; HI, homoplasy index.
Molecular phylogenetic tree inferred from a parsimony analysis of β-tubulin sequences of members of the genus Malassezia. MP bootstrap values >50% in 1000 replications are shown at nodes. The tree is rooted with Cryptococcus neoformans. CI, consistency index; RI, retention index; HI, homoplasy index.
Molecular phylogenetic tree inferred from a parsimony analysis of D1/D2 26S rRNA, ITS-5.8S rRNA and β-tubulin gene sequences of members of the genus Malassezia. MP bootstrap values >50% in 1000 replications are shown at nodes. The tree is rooted with Cryptococcus neoformans. CI, consistency index; RI, retention index; HI, homoplasy index.
The isolates MA 1453 and MA 1456 clustered with M. furfur strains. These isolates had identical D1/D2 and ITS sequences. The similarity of these sequences was 100% between these isolates and M. furfur CBS 7985, which was isolated from an ostrich. A different topology was observed using β-tub sequencing. The isolates MA 1453 and MA 1456 formed a different clade from the M. furfur neotype species.
The isolate MA 1455, belonging to the novel proposed species M. brasiliensis, clustered close to the M. furfur reference strains, but in a different clade with high support in most of the individual gene trees (D1/D2: 67%; ITS: 99%; β-tub: 98%). Dissimilarities between this isolate and M. furfur CBS 1878NT and M. furfur CBS 7019NT in D1/D2 sequences were 1.4% and 1%, respectively. Besides, dissimilarities between MA 1455 and M. furfur CBS 1878NT and M. furfur CBS 7019NT in ITS and β-tub sequences were 4.8% and 5.3%, and 4.3% and 4.5%, respectively.
The isolate MA 1454, belonging to the novel proposed species M. psittaci, clustered close to the type strain of Malassezia yamatoensis, but in a different clade with high support (100%) in all the individual gene trees. The dissimilarity between this isolate and M. yamatoensis CBS 9725T in D1/D2 sequences was 1.7%. Besides, dissimilarities between MA 1454 and M. yamatoensis CBS 9725T in ITS and β-tub sequences were 3.6% and 5.9%, respectively.
Both M. brasiliensis and M. psittaci received strong support (100% bootstrap value) as distinct clades in the combined D1/D2-ITS-β-tub MP tree. They were clearly separated from M. furfur and M. yamatoensis, respectively.
Description of the proposed new speciesM. brasiliensisCabañes, Coutinho, Bragulat et Castellá, sp. nov.
MycoBank MB 812624
M. brasiliensis (brasiliensis: bra.si.li.en'sis M.L. adj, brasiliensis pertaining to Brazil, South America; this Latin-derived species epithet refers to the country from which the yeast was first isolated).
On mDA, after 7 days at 32°C, colonies are large (average diameter 2.5mm, 1.6–3.2mm), whitish to cream-colored, smooth, dull, butyrous, moderately convex and slightly elevated in the center with entire margins. Cells are ovoidal to ellipsoidal, 2.8–3.9μm×1.9–3.3μm, with buds formed monopolarly on a broad base. No growth is obtained on SGA. Catalase reaction is positive, and β-glucosidase activity is negative. Growth occurs on glucose–peptone agar with Tween-20, Tween-40, Tween-60, Tween-80 and Cremophor EL as sole source of lipid. Good growth appears at 37°C and 40°C and no growth occurs at 45°C. The teleomorph is unknown.
The type strain CBS 14135 (=CECT 13138; originally strain MA 1455) was isolated from lesions on the beak of a parrot (A. aestiva) in São Paulo, Brazil. The strains were deposited in the CBS Fungal Biodiversity Centre, Utrecht, The Netherlands, and in the Spanish Type Culture Collection, Valencia, Spain, as CBS 14135 and CECT 13138, respectively.
M. psittaciCabañes, Coutinho, Bragulat et Castellá, sp. nov.
MycoBank MB 812625
M. psittaci (psittaci: psit’ta.ci. L. n. N. psittacus a parrot; M.L. gen. n. psittaci of a parrot; this Latin-derived species epithet refers to the host animal from which the yeast was first isolated).
On mDA, after 7 days at 32°C, colonies are large (average diameter 2.6mm, 1.9–3mm), whitish to cream-colored, smooth, shiny, butyrous, moderately convex and slightly elevated in the center with entire margins. Cells are globose to ovoidal, 2.5–4.4μm×2.2–3.4μm, with buds formed monopolarly on a broad base. No growth is obtained on SGA. Catalase reaction is positive, and β-glucosidase activity is negative. Growth occurs on glucose–peptone agar with Tween-20, Tween-40, Tween-60, Tween-80 and Cremophor EL as sole source of lipid. No growth appears at 37°C. The teleomorph is unknown.
The type strain CBS 14136 (=CECT 13137; originally strain MA 1454) was isolated from lesions on the beak of a parrot (P. menstruus) in São Paulo, Brazil. The strains were deposited in the CBS Fungal Biodiversity Centre, Utrecht, The Netherlands, and in the Spanish Type Culture Collection, Valencia, Spain, as CBS 14136 and CECT 13137, respectively.
DiscussionThe number of recognized Malassezia species described in the present century (n=7) is the same that the number of recognized Malassezia species described in the two last centuries (n=7). In fact, with the exception of M. furfur (Robin) Baillon (1889) and M. pachydermatis (Weidman) Dodge (1925), most of the species were described in the last decade of the 20th century on the basis of morphological, physiological and molecular studies which were already available for these purposes.13,28
Lipid-dependent Malassezia yeasts require complex media enriched with lipids. These particular requirements (e.g. ability to use Tweens 20, 40, 60 and 80) were found to be useful to separate Malassezia species.13,16 However, the addition of the last new species into this genus have resulted in similar physiological patterns among several species, and thus in a doubtful identification. In these cases, the identification should be confirmed by DNA sequencing analysis (e.g. D1/D2 and ITS).14
Malassezia yeasts have been isolated from almost all domestic animals, different wild animals held in captivity, and also from wildlife.29 Despite this, the occurrence of Malassezia yeasts on the skin of most animals remains unknown. Besides, in a recent study of the occurrence of fungal species in human skin,12 DNA sequences that may represent unidentified Malassezia species were also detected. These facts made it possible to anticipate an increase in the number of new species in this genus, particularly if wild animals are studied further.5
This increase may difficult the description of future new species in this genus if we only use the classical techniques of phenotipical characterization (e.g. ability to use Tweens 20, 40, 60 and 80) and the present molecular “gold standard” D1/D2 gene to determine Malassezia species. Various molecular markers had been used to achieve discrimination among the recognized species of Malassezia. The most studied loci had been the D1/D2 and the ITS. In addition to the sequencing of the rRNA genes, other genes such as the chitin synthase-2 gene (CHS2)20 and the β-tub gene6 have been proposed for taxonomic purposes in this genus.
In other fungal genera, such as Penicillium and Aspergillus, the molecular “gold standard” ITS gene does not always distinguish different species. Protein coding genes are widely used in mycology for the identification and have generally a higher interspecies variability than the ITS region. There is no standard choice of a protein-coding gene in the fungal kingdom, but β-tub and calmodulin sequences are frequently used for the identification of Aspergillus and Penicillium species and are better species markers than ITS.19 On the other hand, new tools such as MALDI-TOF mass spectrometry,21 may be useful for a proper characterization of future new species in the genus Malassezia.
The isolates MA 1453 and MA 1456 had both very similar physiological patterns and D1/D2 sequences to M. furfur. However, they form a different clade from the M. furfur neotype strains using both the β-tub sequencing and the multilocus analysis. Only a few strains of M. furfur from birds have been studied in depth. On the other hand, a considerable genetic diversity within this species has been observed. Four distinct LSU rRNA sequences have been reported in this species.15 Three isolates from ostriches (e.g. M. furfur JG 590=CBS 7985) showed the most common LSU rRNA sequence (sequence “a”) together with other human and animal isolates. Two isolates from an elephant (JG 570=CBS 7984) and a pelican (JG 593) clusterized together and showed a unique LSU rRNA sequence (sequence “b”). Eight well-distinguished subtypes in this species were revealed using AFLP.17 Isolates from animals clustered in two subtypes. Subtype 2 contained two human isolates and three isolates from animals (e.g. elephant, elk) and subtype 8 only two isolates from an elephant (JG 570=CBS 7984) and an ostrich (JG 590=CBS 7985).
In our study, the topology of some species was not always concordant between the three markers investigated. This has been also described in other studies in Malassezia species.3,6,14,20 Some species tend to jump between clades depending on the gene analyzed and the set of other fungi included. This implies that further phylogenetic research is needed to establish the position of these species in the tree of life.14 A further explanation of these incongruent results may be that hybridization may have occurred during speciation, and cell fusion, karyogamy and meiosis may be possible within the genus.3 In the case of speciation through clonal divergence and genetic drift, probably followed by some host adaptation, one would expect concordance between the phylogenetic patterns of each individual gene. So, the lack of concordance may indicate that recombination has played a role in the divergence of these species. This is particularly interesting, as sexual reproduction is unknown in Malassezia. Recently, a region corresponding to the mating type locus (MAT) has been identified for these yeasts.26
On the other hand, in the present investigation we propose two new Malassezia species. Among other differences, the new species M. brasiliensis can be distinguished from M. pachydermatis by its inability to grow in SGA, from Malassezia caprae, Malassezia dermatis, Malassezia equina, Malassezia japonica, Malassezia nana, M. psittaci and M. yamatoensis by its ability to grow at 40°C, from Malassezia cuniculi, Malassezia globosa and Malassezia obtusa by its ability to assimilate Tween-20, Tween-40, Tween-60 and Tween-80, from Malassezia slooffiae and M. sympodialis by its ability to assimilate Cremophor EL, from Malassezia restricta by its catalase activity, and from M. furfur by its β-glucosidase activity. The new species M. psittaci can be distinguished from M. pachydermatis by its inability to grow in SGA, from M. brasiliensis, M. dermatis, M. furfur, M. japonica, M. nana, M. slooffiae, M. sympodialis and M. yamatoensis by its inability to grow at 37°C, from M. cuniculi, M. globosa and M. obtusa by its ability to assimilate Tween-20, Tween-40, Tween-60 and Tween-80, from M. caprae and M. equina by its ability to assimilate Cremophor EL, and from M. restricta by its catalase activity.
In addition, the validation of these new species was supported by the analysis of the D1/D2, the ITS and the β-tub genes which confirmed the separation of these new species from the other described species of the genus. Although a boundary between species (a prerequisite number of nucleotide differences in either regions to separate species) has been not clearly stated, the within-species D1/D2 sequence similarity is above 99% for Malassezia species (<1% of dissimilarity).17,29 In our study, the multilocus sequence analysis of the three loci provides also robust support to delineate these new species. So, phylogenetic analysis of sequences from M. brasiliensis and M. psittaci showed that they were clearly distinct from the other 14 described Malassezia species, exceeding the variation generally observed to occur between basidiomycetous yeast species.27 On the other hand, our study confirms also the presence of lipid-dependent Malassezia yeasts on parrots.
Conflict of interestThe authors have no conflict of interest.
The authors thank Lab&Vet Diagnóstico e Consultoria Veterinària for kindly providing the parrot samples and Suzana Maria Bezerra from the Laboratory of Molecular and Cellular Biology of the Paulista University (PU) and Carolina Gómez from the Veterinary Mycology Group of the Universitat Autònoma de Barcelona (UAB) for their valuable technical assistance. Financial support came from Servei Veterinari de Bacteriologia i Micologia of the UAB and from the Research Section of the PU.