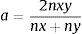Over the last decades, Candida species have emerged as important pathogens in immunocompromised patients. Nosocomial infections are mainly of endogenous origin. Nevertheless, some cases of exogenous candidiasis have also been reported.
AimsThe aim of this study was to evaluate the genetic relatedness between Candida albicans, Candida glabrata, Candida tropicalis, Candida krusei and Candida kefyr isolates recovered from intensive care unit (ICU) patients.
MethodsA total of 132 Candida clinical isolates (62 C. albicans, 40 C. glabrata, 13 C. tropicalis, 11 C. krusei, 6 C. kefyr), obtained from specimens of endotracheal aspirate, urine and blood taken from patients of a tertiary hospital in Poland, were included in the study. Species identification was performed by PCR method and genetic relatedness was assessed by randomly amplified polymorphic DNA assay (RAPD) with five primers.
ResultsThe RAPD analysis revealed high genetic diversity among the studied Candida isolates, indicating that most of the strains were from endogenous sources. Only two clonal strains of C. glabrata isolated from different patients were observed, suggesting a possible cross-transmission of these pathogens.
ConclusionsOur study confirmed the high discriminatory power of the RAPD assay. This genotyping method can be applied to local epidemiological studies of Candida species.
En las últimas décadas, el hongo Candida se ha convertido en un patógeno importante para los pacientes con trastornos del sistema inmune. Las infecciones nosocomiales son fundamentalmente de origen endógeno; sin embargo, también se han documentado algunos casos de candidiasis exógena.
ObjetivosEl objetivo del estudio fue evaluar la relación genética entre las cepas de Candida albicans, Candida glabrata, Candida tropicalis, Candida krusei y Candida kefyr aisladas de pacientes en cuidados intensivos.
MétodosSe estudiaron 132 aislamientos de Candida (62 C. albicans, 40 C. glabrata, 13 C. tropicalis, 11 C. krusei, 6 C. kefyr) obtenidos de muestras procedentes de aspirado endotraqueal, orina y sangre tomadas de pacientes de un hospital en Polonia. La identificación de las especies se realizó mediante PCR, y el estudio de la relación genética con el método de amplificación aleatoria de ADN polimórfico (RAPD) con cinco oligonucleótidos.
ResultadosEl análisis de la amplificación por RAPD mostró una alta diversidad genética entre los aislamientos objeto de estudio, lo que indica que la mayoría de ellos tenían un origen endógeno. Solo se observaron dos cepas clonales de C. glabrata procedentes de diferentes pacientes, lo que evidencia una posible transmisión cruzada de estos patógenos.
ConclusionesNuestro estudio confirma el alto poder discriminatorio de la técnica RAPD, lo que validaría este método de genotipificación para el estudio de la epidemiología local de especies de Candida.
In the last decades Candida species have emerged as important nosocomial pathogens in immunocompromised individuals with severe underlying diseases and comorbidities, such as patients treated in intensive care units (ICU).11,23
Candida resides as a commensal in the oral cavity, gastrointestinal tract and vagina of healthy humans. Due to prior colonization, nosocomial infections caused by these yeasts are considered mainly endogenous. Nevertheless, some cases of exogenous candidiasis, occurring as a result of dissemination of Candida species in the hospital environment, have also been reported.3,5
Candida albicans remains the predominant species recovered from clinical specimens worldwide. However, in the last two decades other species of this genus have become increasingly prevalent. The distribution of Candida varies according to the geographic areas. The most commonly observed non-C. albicans Candida species in studies from Central and Northern Europe is Candida glabrata.2,8,15,26,34 The other species are observed less frequently, although it has been reported that infections due to Candida tropicalis have increased on a global scale.20 Also the frequency of isolation of Candida krusei in Poland and other several Eastern European countries is much higher than in other geographic areas.27 Furthermore, new emerging pathogenic Candida species have been described: Candida dubliniensis and Candida africana, phenotypically similar to C. albicans, as well as Candida nivariensis and Candida bracarensis, closely related to C. glabrata. These pathogens can be easily misidentified by conventional diagnostic methods, because there is only little evidence for differences between their phenotypes.12
In most cases, Candida identification to the species level allows to predict their drug susceptibility. There is no transfer of resistance genes between yeasts. The acquisition of resistance is observed mainly in restricted clinical settings, due to prolonged antifungal treatment. Different susceptibility patterns to antifungal agents among the species is of particular concern. The increase of non-C. albicans Candida species isolation is accompanied by a higher proportion of isolates, which are frequently resistant to fluconazole. C. krusei has an inherited resistance to fluconazole and has been recognized as a potentially multidrug-resistant (MDR) fungal pathogen, due to its decreased susceptibility to both 5-fluorocytosine and amphotericin B.27C. glabrata isolates are also more resistant to fluconazole.14 Several identified strains of C. nivariensis and C. bracarensis were resistant to azoles or amphotericin B or exhibited a decreased susceptibility to these antimicrobials.6,16
Understanding the local epidemiology of Candida is of great relevance for the clinical management and treatment of candidiasis, especially in critically ill patients. Therefore, it is important to determine the prevalence and diversity of Candida, as well as the phenotypic and genotypic characteristics of these pathogens.10
From a variety of molecular techniques, PCR-based approaches for molecular typing are applied with most success. They are simple, as well as rapid and cost-effective. The above-mentioned advantages of PCR techniques and their accuracy make them suitable tools for epidemiological investigation of Candida species on both global and local levels.1 The aim of this study was to evaluate the genetic relatedness among selected Candida isolates, recovered from the ICU patients of a tertiary hospital in Krakow, Poland over a 4-year period, by randomly amplified polymorphic DNA assay (RAPD).
Material and methodsPatients and strainsOne hundred and twenty three patients with multi-organ failure were enrolled into the study. They were hospitalised between 2009 and 2012 at the Intensive Care Unit of Ludwik Rydygier Memorial Hospital in Krakow. The study population consisted of 74 (60.2%) males and 49 (39.8%) females with the average age of 64 years (range 18–93) from south-eastern Poland. Antifungal prophylaxis had not been administered to the patients included in this study.
The predominant fungal species isolated in the study period were C. albicans (52.6%) and C. glabrata (16.2%), followed by C. tropicalis (6.7%), C. krusei (5.3%) and Candida kefyr (3.1%), whereas other Candida species were isolated less frequently (<2%). For the genotyping analysis we selected approximately 40% of all C. albicans, C. glabrata, C. tropicalis, C. krusei and C. kefyr isolates recovered from endotracheal aspirate (ETA), urine and blood from ICU patients of a Krakow hospital in the years 2009–2012 (Table 1).
Distribution of the Candida species isolated from ICU patients of Ludwik Rydygier Memorial Hospital in Krakow from ETA and urine (at concentration ≥104CFU/ml) and blood samples, between 2009 and 2012.
| Species | Total no. of strains isolated between 2009 and 2012 | No. (%) of strains included into the molecular studies |
|---|---|---|
| C. albicans | 169 | 62 (36.7) |
| C. glabrata | 74 | 40 (54.1) |
| C. tropicalis | 41 | 13 (31.7) |
| C. krusei | 29 | 11 (37.9) |
| C. kefyr | 15 | 6 (40.0) |
| Total | 328 | 132 (40.2) |
A total of 132 clinical isolates (62 C. albicans, 40 C. glabrata, 13 C. tropicalis, 11 C. krusei and 6 C. kefyr isolates) were included in the study. One hundred and twenty Candida isolates (55 C. albicans, 36 C. glabrata, 13 C. tropicalis, 11 C. krusei and 5 C. kefyr) were obtained from ETA, while nine (6 C. albicans and 3 C. glabrata) were taken from urine. These isolates were cultured at a concentration ≥104CFU/ml, indicating that the fungal colonization was high. Furthermore, one isolate each of the species C. albicans, C. glabrata and C. kefyr were recovered from blood. Each isolate was obtained from different patients, with the following exceptions: C. albicans CA1 and CA3, as well as CA46 and CA47 strains, were acquired from ETA and urine of one subject. Both C. albicans and one isolate each of the species C. glabrata, C tropicalis, C. krusei and C. kefyr were cultured from seven of the studied individuals.
The study was approved by the Ethics Committee of Jagiellonian University Medical College (KBET/129/B/2011).
Phenotypic characterization of strainsEndotracheal aspirate and urine specimens were cultured by quantitative technique on Sabouraud glucose agar (SGA) (bioMérieux, Marcy l’Etoile, France) and incubated at 37°C for 48h. Fungal concentration of the samples were inferred between 104CFU/ml and 106CFU/ml after 48h of incubation.24 The yeasts recovered from blood were detected in cultures using the automated BacT/ALERT system (bioMérieux) as a routine hospital practice. For further identification, they were plated and cultured on SGA.
The species identification was based on the colony color of the isolates on chromID Candida medium (bioMérieux) and the analysis of the biochemical profiles was made using VITEK 2 Compact with YST ID card (bioMérieux). Additionally, molecular identification of C. albicans and C. glabrata isolates was performed.
In vitro susceptibility of the isolates to several antifungal agents (amphotericin B, fluconazole, 5-fluorocytosine and voriconazole) was analyzed with VITEK 2 Compact system (card number AST-YS01), according to the M27-A3 guidelines of Clinical and Laboratory Standards Institute criteria.9 The reference strains included C. albicans ATCC 90028 and C. glabrata ATCC 2001.
Molecular characterization of strainsMolecular tests were conducted in the Department of Pharmaceutical Microbiology Jagiellonian University Medical College. Fungal genomic DNA was extracted with Genomic Mini AX YEAST isolation kit (A&A Biotechnology, Gdynia, Poland), according to the manufacturer's instructions. The fungal isolates were identified and genotyped by PCR analysis. The PCR reactions were performed in a volume of 25μl containing the following: 1×PCR buffer, 0.2mM each of dATP, dGTP, dCTP, and dTTP (Promega, Madison, Wisconsin, USA), 0.4μM of each of the primers (Blirt, Gdańsk, Poland), 25ng of genomic DNA, 2mM MgCl2, and 0.625U of Taq DNA polymerase (Promega, Madison, Wisconsin, USA). Amplification was carried out in a T personal thermocycler (Biometra, Göttingen, Germany). The DNA fragments were separated by 2% agarose gel electrophoresis and visualized with ethidium bromide staining (Sigma-Aldrich Chemie, Munich, Germany). The PCR product size was determined by comparison with the molecular weight standard (O’Gene Ruler 100bp DNA Ladder Plus; Thermo Fisher Scientific, Waltham, Massachusetts, USA).
The molecular identification of C. albicans was carried out by PCR assay with CR-f (5′–GCTACCACTTCAGAATCATCATC–3′) and CR-r (5′–GCACCTTCAGTCGTAGAGACG–3′) species-specific primers, targeting the HWP1 gene, as previously described.28 The PCR conditions were as follows: denaturation at 95°C for 2min, 30 cycles of denaturation at 94°C for 45s, annealing at 58°C for 40s, and extension at 72°C for 55s, followed by a final extension at 72°C for 10min.
Molecular confirmation of C. glabrata species was performed by PCR reaction with CGL1 (5′–TTATCACACGACTCGACACT–3′) and CGL2 (5′–CCCACATACTGATATGGCCTACAA–3′) primers, designed by Luo and Mitchell, based on the sequence data for the internal transcribed spacer (ITS) region.21 Amplification was carried out with an initial denaturation at 95°C for 2min, followed by 40 cycles consisting of denaturation at 94°C for 30s, annealing at 58°C for 30s, and elongation at 72°C for 30s, and a final extension at 72°C for 10min.
The genetic relatedness among the clinical C. albicans and C. glabrata isolates was characterized by RAPD assay. Five arbitrary primers, CD16AS, HP1247, ERIC-2, OPE-3, and OPE-18 (Genomed, Warszawa, Poland), previously applied in our study,25 were used in the analysis. Due to the low discriminatory power of OPE-18, we have excluded this primer from the analysis of genetic relatedness among C. tropicalis, C. krusei and C. kefyr isolates. PCR was conducted with the following cycle profile: one cycle at 95°C for 2min, followed by 40 cycles at 95°C for 1min, 36°C for 1min, 72°C for 2min, and a final extension at 72°C for 5min. Similarity analysis of RAPD patterns was carried out with the BIO 1D++ software (Vilber-Lourmat, Marne La Vallée, France). Therefore, the digital images of RAPD patterns, obtained with all the primers tested, were used for comparison and clustering. In each sample the presence (1) or absence (0) of RAPD bands was scored in a binary matrix. Those data were stored in a built-in database of the software. Comparison of the binary data was performed via unweighted pair-group method with arithmetic averages (UPGMA). The binary matrix was translated into a distance matrix, using a Dice similarity coefficient. The general formula of the Dice coefficient is as follows:
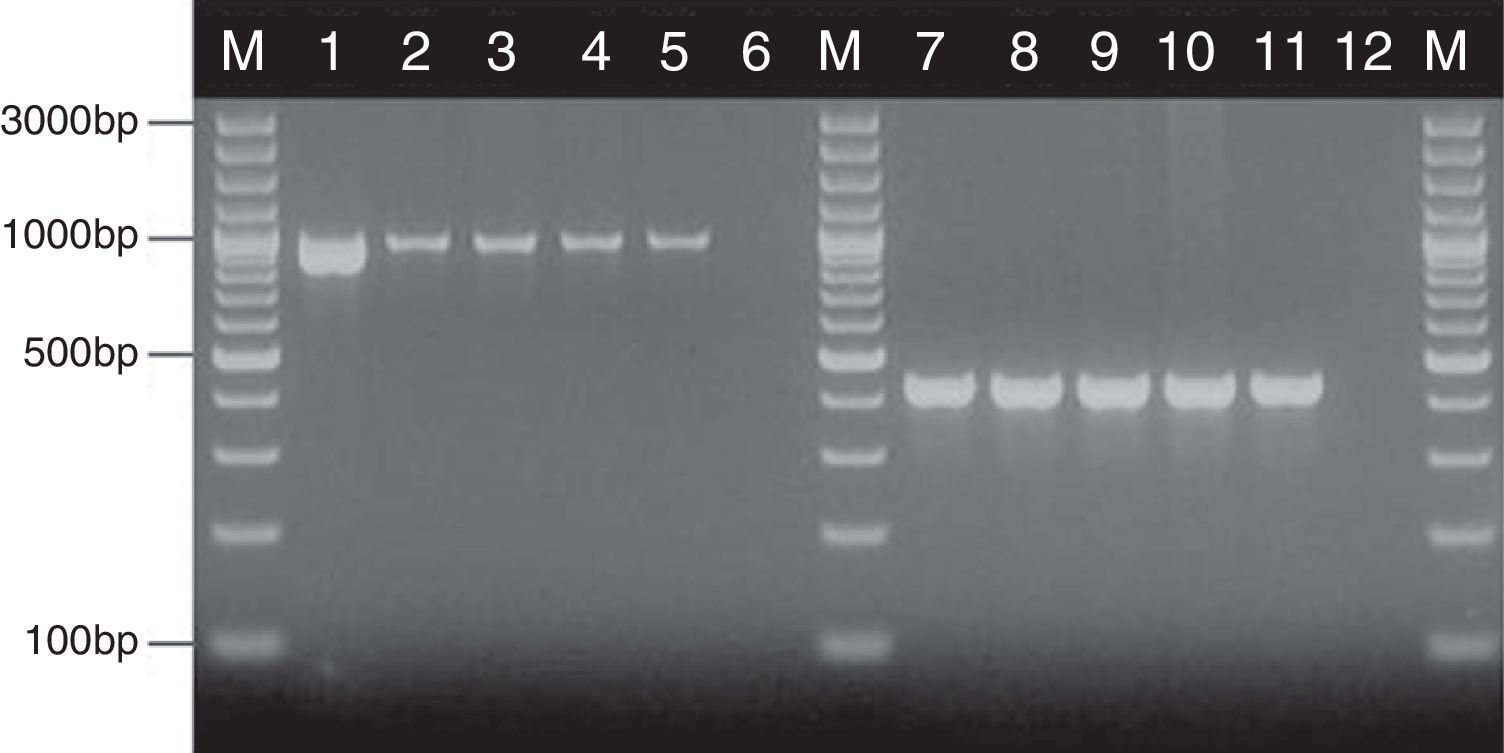
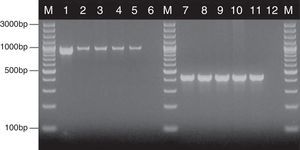
where nx and ny are the number of bands in the lane ‘x’ and in the lane ‘y’ respectively, and nxy the number of the shared bands between the 2 lanes. As a result of the hierarchical cluster analysis by UPGMA, composite dendrograms for each Candida species were constructed. Based on the similarity value, the strains were determined as identical (100%), genetically related (80–99%) or unrelated (<80%). The experiments were carried out in duplicates and there were no differences in the RAPD profiles.ResultsAll the 62 isolates subjected to PCR assay using primers targeting the HWP1 gene were identified as C. albicans, as a ∼1000-bp fragment was amplified. In the remaining 40 isolates, analyzed with the use of the primers for amplification of the ITS region, a ∼420-bp band, specific for C. glabrata was detected (Fig. 1).
Detection of genes encoding HWP1 protein and the ITS region, by PCR method among selected C. albicans and C. glabrata isolates from ICU. Lane M: 100-bp molecular size marker (Thermo Fisher Scientific, Waltham, Massachusetts, USA); lane 1: C. albicans ATCC 90028; clinical C. albicans strains in lane 2: CA1; lane 3: CA22; lane 4: CA39; lane 5: CA48. Lane 7: C. glabrata ATCC 2001; clinical C. glabrata strains in lane 8: CG1; lane 9: CG12; lane 10: CG24; lane 11: CG31. Lanes 6 and 12: negative controls.
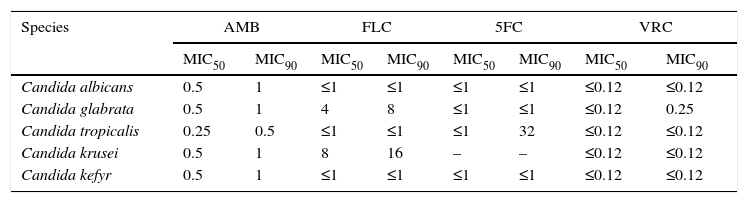
The MIC50 and MIC90 values of amphotericin B, fluconazole, 5-fluorocytosine and voriconazole for C. albicans, C. glabrata, C. tropicalis, C. krusei and C. kefyr isolates are shown in Table 2. All 102 Candida isolates were susceptible to the mentioned antifungal agents, except for two C. glabrata strains (CG14 and CG30), that showed dose-dependent susceptibility to fluconazole (MIC 16 and 32mg/l) and two C. tropicalis strains with intermediate susceptibility to fluconazole (CT10 and CT13) (MIC 32mg/l).
The MIC50 and MIC90 values [mg/l] of antifugal agents for Candida albicans, Candida glabrata, Candida tropicalis, Candida krusei and Candida kefyr isolates.
| Species | AMB | FLC | 5FC | VRC | ||||
|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| Candida albicans | 0.5 | 1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤0.12 | ≤0.12 |
| Candida glabrata | 0.5 | 1 | 4 | 8 | ≤1 | ≤1 | ≤0.12 | 0.25 |
| Candida tropicalis | 0.25 | 0.5 | ≤1 | ≤1 | ≤1 | 32 | ≤0.12 | ≤0.12 |
| Candida krusei | 0.5 | 1 | 8 | 16 | – | – | ≤0.12 | ≤0.12 |
| Candida kefyr | 0.5 | 1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤0.12 | ≤0.12 |
AMB, amphotericin B; FLC, fluconazole; 5FC, 5-fluorocytosine; VRC, voriconazole.
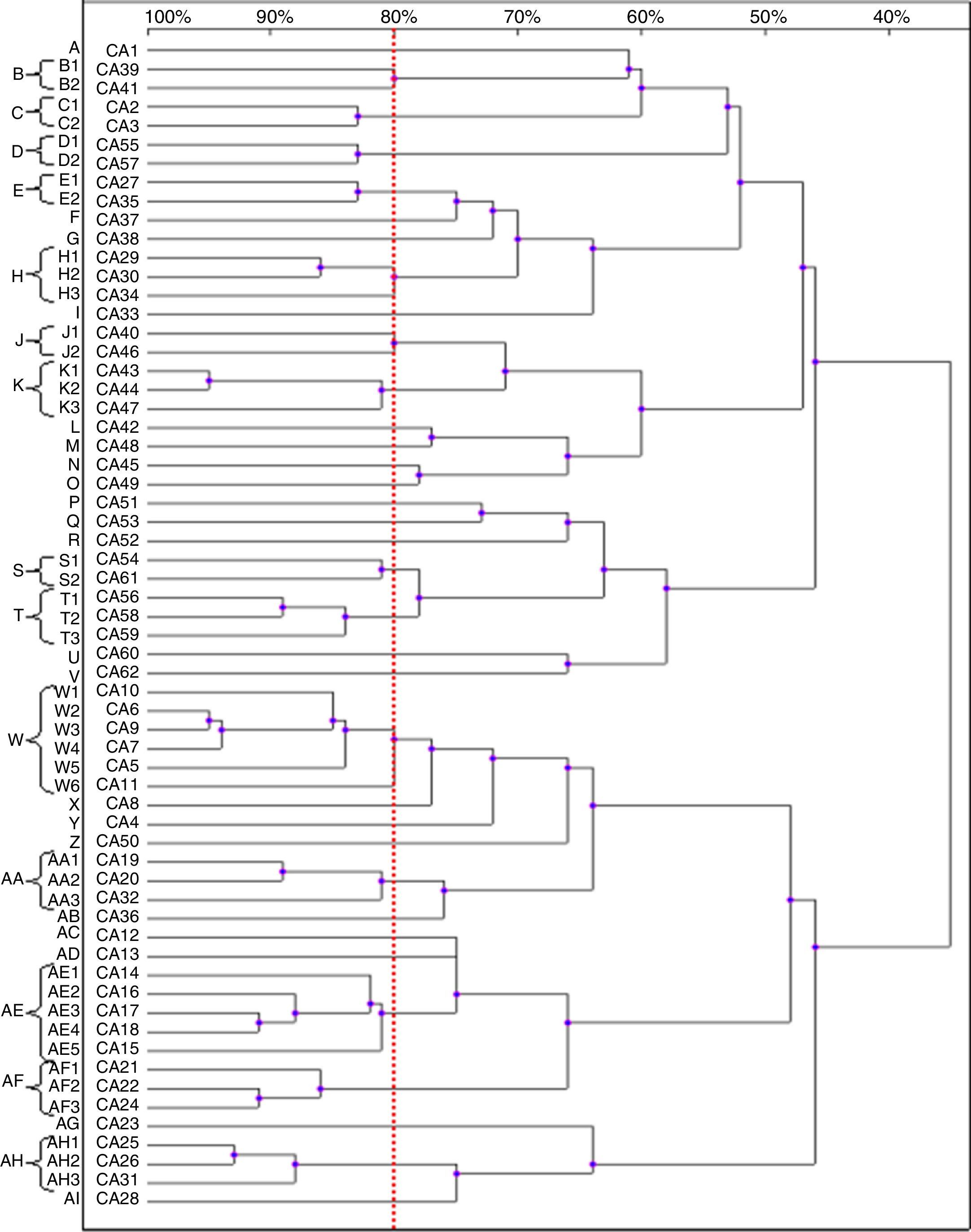
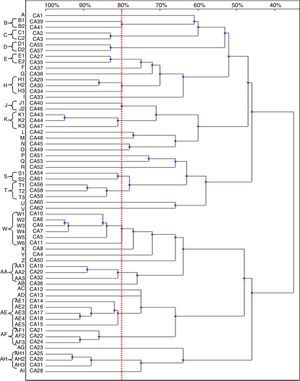
Based on the results of the RAPD reactions with all the primers tested, we constructed the dendrograms. Among the 62 C. albicans isolates, 62 RAPD profiles were distinguished. A total of 41 C. albicans isolates with a similarity level higher than 80% were allocated into 14 clusters (B–E, H, J, K, S, T, W, AA, AE, AF and AH). The remaining 21 strains, which presented a unique pattern each (with similarity level less than 80%), were considered as unrelated (A, F, G, I, L–R, U, V, X–Z, AB, AC, AD, AG and AJ). The dendrogram distinguished clusters W and AE, that consisted of six and five strains, respectively. Furthermore, we observed six groups (H, K, T, AA, AF and AH) containing three strains each, while clusters B–E, J and S were shared by two strains each (Fig. 2).
Most of the genetically related isolates were recovered from different ICU patients in the same year, with four months as the average time interval of isolation. We did not observe homology between the C. albicans strains obtained from different body sites of the same subjects (isolates CA1 and CA3, as well as CA46 and CA47).
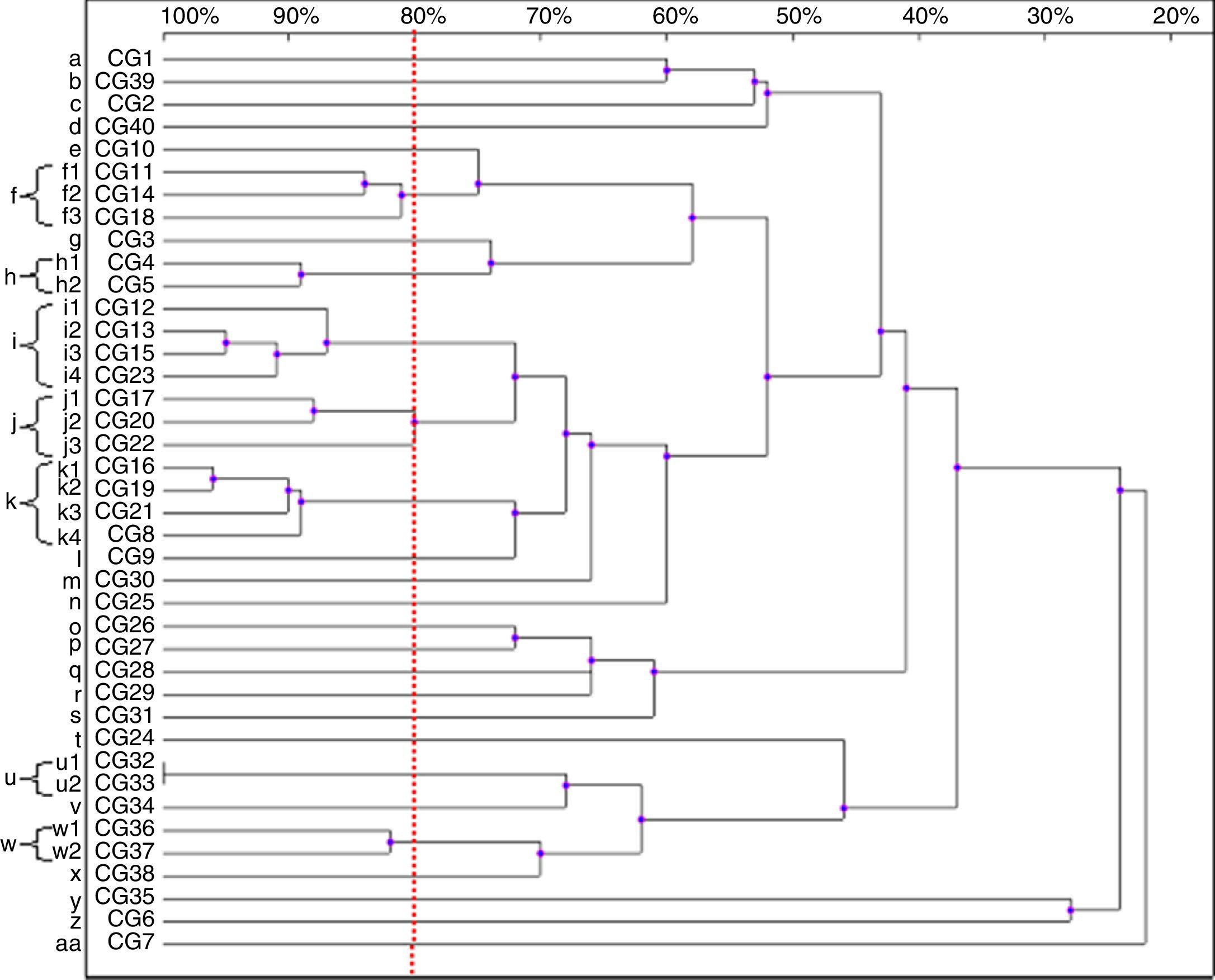
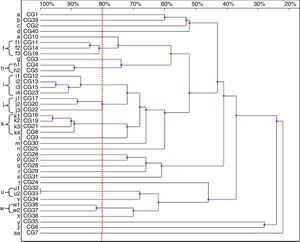
Thirty-nine patterns were observed in the RAPD analysis of the 40 C. glabrata isolates. The strains were allocated into seven clusters (f, h–k, u and w) with similarity values above 80%. The other 22 genotypes (a–e, g, l–t, v, x–z and aa) were represented by single isolates only (Fig. 3).
Four isolates belonged to the clusters ‘i’ and ‘k’. The clusters ‘f’ and ‘j’ consisted of three strains each, while the remaining groups, ‘h’, ‘u’ and ‘w’, included two isolates each. Among the C. glabrata strains, two isolates, CG32 and CG33 (cluster ‘u’), presented 100% homology. They were collected at an interval of two weeks from two different patients hospitalised in the ICU.
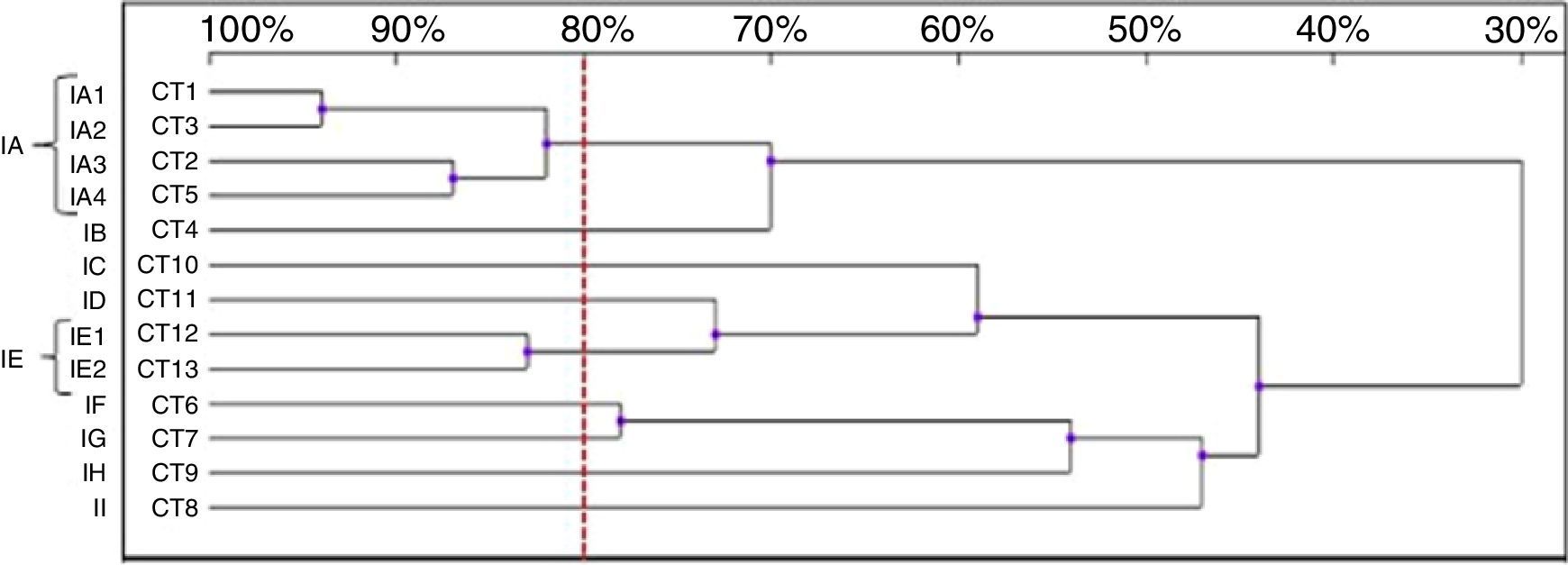
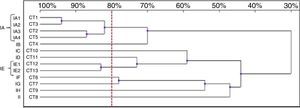
Thirteen RAPD profiles were found in the analysis of the 13 C. tropicalis isolates. The dendrogram allowed to distinguish two clusters (IA – four isolates, IE – two isolates), including isolates that presented similarity values above 80%. The remaining seven strains (IB-ID, IG-II) showed unique patterns (Fig. 4).
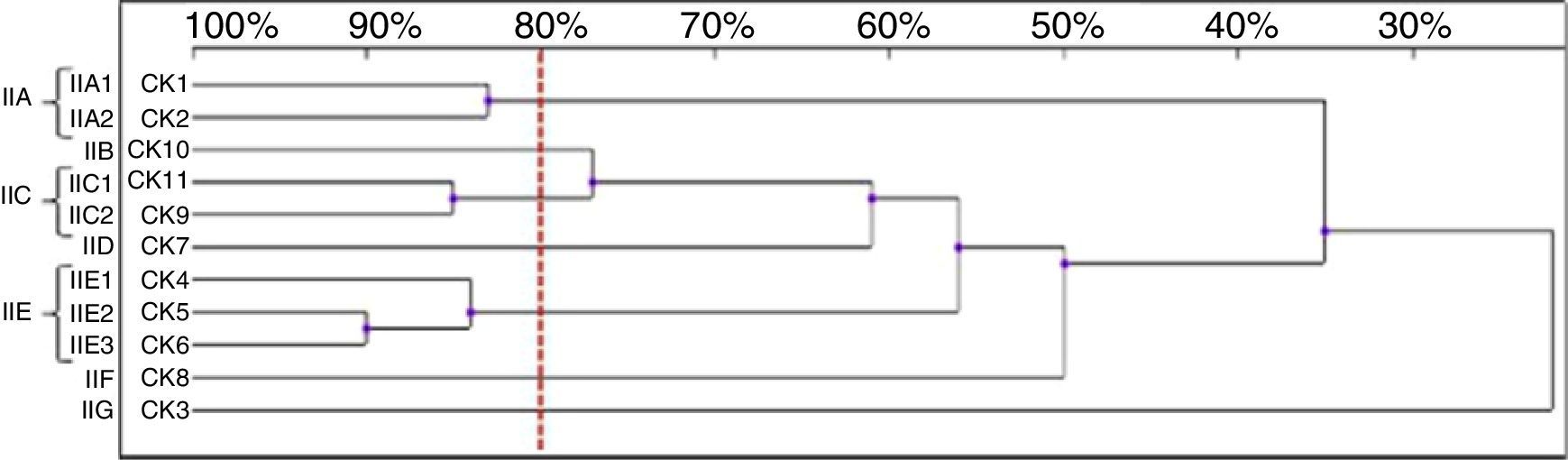
Among the 11 C. krusei strains, 11 RAPD profiles were distinguished. Seven isolates, with similarity values higher than 80%, were allocated into three clusters (IIA, IIC and IIE), whereas four isolates were considered as unrelated (IIB, IID, IIF, IIG) (Fig. 5).
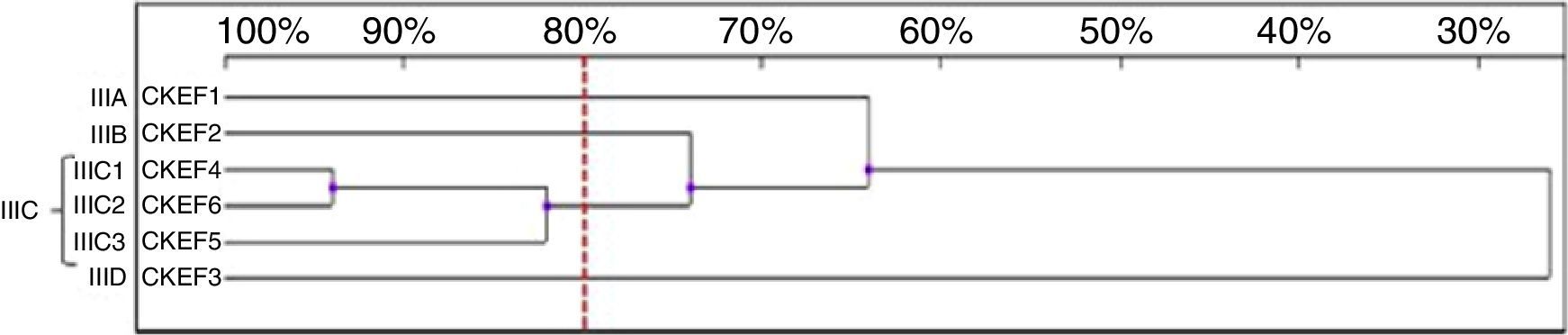
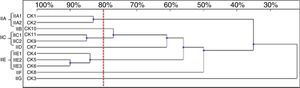
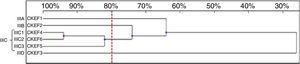
Six genotypes were observed in the RAPD analysis of the six C. kefyr isolates. The dendrogram enable to distinguish three related strains allocated into cluster IIIC. The other three isolates (IIIA, IIIB, IIID) were considered as unrelated (Fig. 6).
The discriminatory power of the RAPD technique was quantified for each species using the Simpson's index.18 In our study the discriminatory power value was 1 for C. albicans, C. tropicalis, C. krusei, and C. kefyr, and 0.99 for C. glabrata.
DiscussionThe yeasts belonging to the Candida genus are recognized as the dominant human pathogenic fungi. They affect hospitalized patients with systemic life-threatening infections.
Because of a documented increase in the incidence of new emerging species closely related to C. albicans and C. glabrata, application of PCR-based molecular methods to identify them is crucial.6,7,32,33 The identification by phenotypic methods (e.g. chromogenic media or automatic systems) may be inconclusive, therefore strain genotyping without a previous molecular identification of the species can be misleading. Using the PCR technique we identified each of the clinical isolates as either C. albicans or C. glabrata.
The accurate identification and molecular typing of Candida species provide valuable information supplementing the growing body of knowledge in the field of their epidemiology. This is of great importance for prevention and control of nosocomial infections caused by these yeasts. The presented findings confirmed the suitability and high discriminatory power of RAPD in the genotyping of Candida isolates on a local level.
The choice of the appropriate method of genotyping depends on the type of the conducted research (e.g. local or global studies, investigation of the origin of infection, studying the routes of transmission of strains, determination of the diversity of isolates), as well as on the financial capabilities and equipment availability. Molecular techniques such as RAPD, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP) and microsatellite typing differ in their discriminatory power, reproducibility, as well as in their performance and results interpretation.
Despite the lack of reproducibility, the RAPD technique is extensively used in local studies because it presents many advantages such as simplicity, rapidity and cost-effectiveness of performing. While single short primers of arbitrary sequence were employed in RAPD, it does not require any DNA sequence information. However, the lack of standardization and difficulties in inter laboratory comparison of the results, make this method not suitable in a global level epidemiological investigation.19,29
One of the techniques utilized for epidemiological investigation of Candida species is RFLP, in which a total genomic DNA is split by a frequent cutting restriction endonuclease. The analysis of the obtained band pattern is essential for the differentiation of the strains. The RFLP is considered an easy, rapid and cost-effective typing method. The low discriminatory power and difficulties in the interpretation of formed results are the major drawbacks of RFLP. Another typing technique, that provides highly resolutive, reliable and reproducible results, was AFLP. However, due to multiple-step procedure, AFLP implies high cost and the necessity of having well-trained personnel, so it has been rarely utilized in routine use. Microsatellite typing is the further technique associated with genetic analysis of fungi, which possess high discriminatory power. While this method is very suitable for application in large-scale epidemiological studies, it is expensive and requires specialized equipment. Besides, researchers have encountered some difficulties when implementing it in the routine use.29
In the current study, most Candida isolates were patient-specific, indicating their endogenous source. Our results are in agreement with the work published by da Costa et al., who studied the genetic relatedness of 15 C. albicans and 15 C. tropicalis strains recovered from whole saliva. The authors did not demonstrate any identical strains among C. albicans isolates, however they found three isolates of C. tropicalis, which presented the same RAPD pattern.13 Marol et al., who studied Candida infections occurring in the anesthesiological intensive care unit of a Turkish hospital, revealed that most of the infections seemed to have an endogenous source. The investigators found only three pairs of C. albicans strains, recovered within short period of time (from one to four days), as well as three C. tropicalis strains, isolated between one and six months, with the same RAPD profiles.22 Similarly, Vrioni and Matsiota-Bernard genotyped 17 C. albicans and 16 C. tropicalis strains isolated from transtracheal aspirate, urine, wound, abscess, and blood samples of ICU patients, and they found two identical C. albicans isolates in urine of two different patients. It is worth noting that the aforementioned C. albicans strains were isolated in a time interval less than one month. The investigators confirmed much lower genetic variability among C. tropicalis strains. They could distinguish only six different RAPD pattern combinations.31 The RAPD analysis of 17 C. kefyr strains performed by Kalkanci et al. revealed that the strains exhibited seven different genotypes. These strains were obtained from different patients with hematological malignancies of the same hospital over an eight month-period.19 These findings, together with the results of the present study, confirm that Candida infection can be spread exogenously. The analysis allowed us to recognize two identical C. glabrata strains. Both of them were isolated from endotracheal aspirate from subjects hospitalised within the same month, and these patients had an overlapping ICU stay. These isolates may come either from other patients flora, medical devices, hospital staff or environment. Further studies with larger numbers of samples from different sources (as described above) are required to identify exogenous routs of transmission.
In our study, the homological strains as well as the most of the genotyped isolates were recovered from endotracheal aspirate. In mechanically ventilated ICU patients the endotracheal tube facilitates the passage of Candida from the mouth into the lower airway and can be a reservoir of the pathogens in ventilator-associated pneumonia patients. These observations are supported by Heo et al., who reported that most of the analyzed oral and tracheo-bronchial strains were genetically indistinguishable within a patient.17 However, when these authors compared strains from different patients, they found two homological genotypes, thus demonstrating a risk of microbial acquisition after admission to the hospital.
In this study it can be observed that in a few groups of Candida strains with similarity values between 80 and 99%, the isolates showed only minor differences in the band patterns. The observed differences may be attributed to microevolutions promoted by prolonged colonization, and their emergence depends on the genetic stability of strains in hospital environment.5,30 However, microevolutionary changes can be established only with more discriminatory molecular methods, including sequencing of the RAPD bands.4
To conclude, RAPD analysis with multiple oligonucleotides presented good discriminatory power and can be applied successfully in the epidemiological studies of genomic variability within the Candida species. To assess a probable relationship between acquired infections and their sources further investigations with additional environmental and clinical samples are required.
Conflict of interestThe authors declare that there are no conflicts of interest.
The scientific work was supported by the Jagiellonian University Medical College with funds from maintenance of the research potential of the JU MC Department of Pharmaceutical Microbiology.