A 27-year-old male rural worker was admitted with a fungal keratitis due to an injury involving plant detritus.
Materials and methodsSpecimens were collected for microscopy examination and culture. The isolate was identified by morphological and molecular criteria. Susceptibility testing was performed using CLSI methods. CYP51A gene was PCR amplified and sequenced.
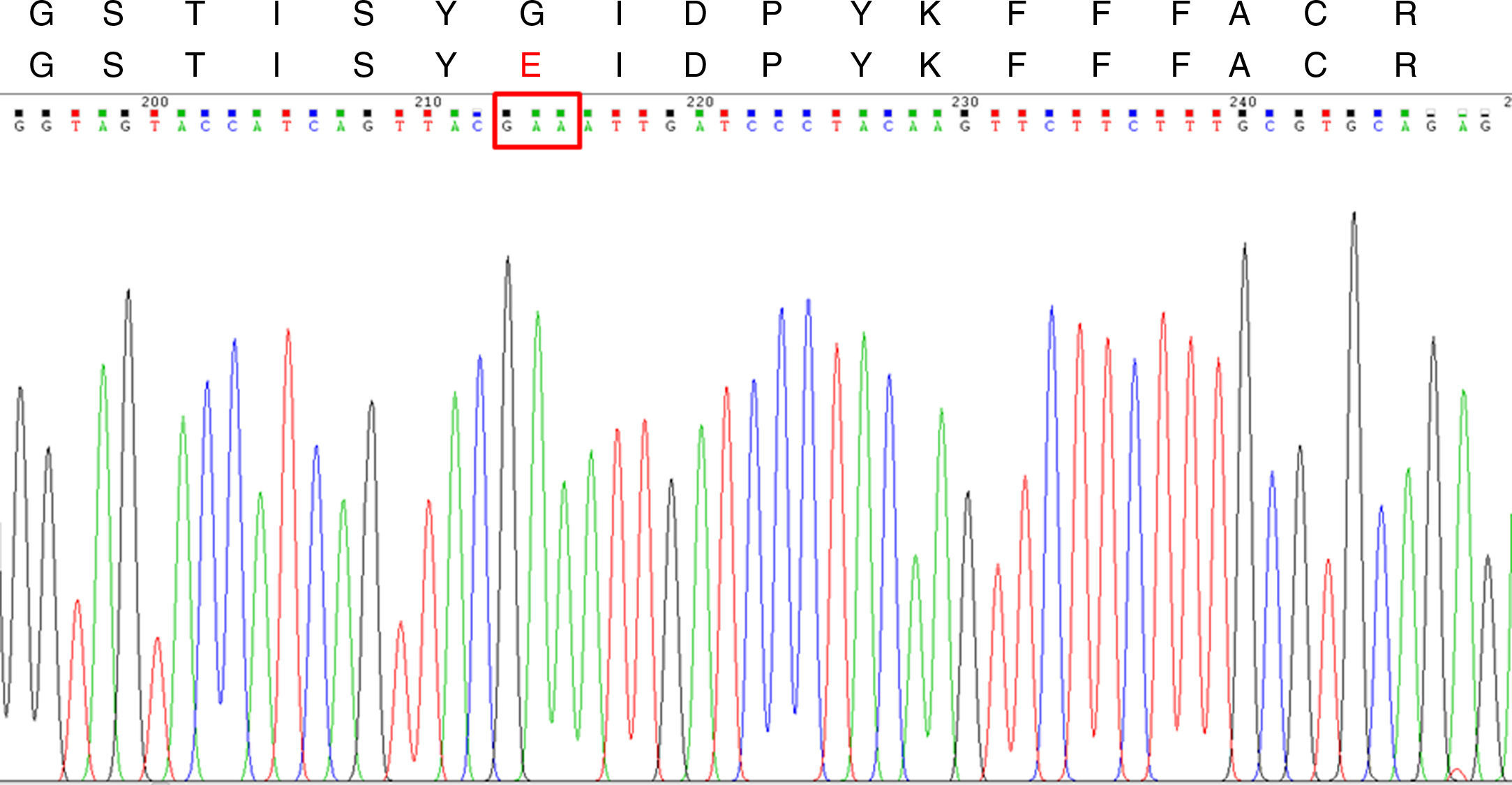
ResultsAn Aspergillus fumigatus strain resistant to itraconazole (MIC>8μg/ml) was isolated. The isolate was susceptible to amphotericin B, posaconazole, voriconazole and caspofungin. CYP51A sequencing showed two mutations leading on the G54E substitution. The patient received natamycin as treatment.
ConclusionsThis is the first report in South America of a clinical A. fumigatus strain carrying the substitution G54E at Cyp51Ap associated with itraconazole resistance. Considering the patient was azole-naive, this resistant isolate may have been acquired from the environment.
Un trabajador rural de 27años de edad fue hospitalizado con una queratitis fúngica debido a un traumatismo con un resto vegetal.
Materiales y métodosSe tomaron las muestras para los exámenes de microscopía y cultivo. El aislamiento se identificó mediante criterios morfológicos y moleculares. Se realizaron pruebas de sensibilidad a los antifúngicos siguiendo el documento del CLSI. Se amplificó y secuenció el gen CYP51A de la cepa.
ResultadosSe aisló una cepa de Aspergillus fumigatus resistente a itraconazol (CIM>8μg/ml). El aislamiento resultó sensible a la anfotericina B, el posaconazol, el voriconazol y la caspofungina. La secuenciación del gen CYP51 reveló 2 mutaciones que generan la sustitución G54E. El paciente fue tratado con natamicina oftálmica.
ConclusionesEste es el primer caso informado en Sudamérica de una cepa clínica de A. fumigatus con la sustitución G54E en el Cyp51Ap, asociada con resistencia al itraconazol. Teniendo en cuenta que el paciente no había recibido nunca antes tratamiento alguno con azoles, podría haber adquirido esta cepa resistente del ambiente.
A 27-year-old male rural worker was admitted to the “Dr. José María Cullen” Hospital (Santa Fe – Argentina) in November 2013 with a corneal ulcer. The patient had suffered an ocular traumatism with a vegetal detritus while he was working in harvesting vegetables. At patient arrival, ocular samples were obtained at the Ophthalmology Department of the hospital. Conjunctival mucopurulent exudate and corneal surface samples were obtained by swabbing and by scrapping with a Kimura spatula, respectively. Samples were immediately sent to the Microbiology Laboratory for their processing. Samples were Giemsa stained and cultured in Sabouraud Dextrose Agar (SDA) with chloramphenicol (Britania, Argentina) and in Thioglycollate broth. Meanwhile, the patient received empirical treatment with oral fluconazole (400mg/day) and ophthalmic natamycin (5% solution), two drops every 30min during the first 24h, together with intravenous ceftazidime and vancomycin. During patient's first day of hospitalization the laboratory informed that septate fungal hyphae were seen in the Giemsa stain, leading to the diagnosis of a keratitis due to a hialohyphomycete. With these data, fluconazole was suspended and natamycin and antibacterials were maintained. Natamycin dosage intervals were changed after the second hospitalization day to two drops every hour during 7 days. Then, it was slowly tapered following the good clinical response until the complete resolution of the infection. After the third hospitalization day, colonies of Aspergillus fumigatus grew in all the SDA slants.
The strain was identified as A. fumigatus by morphological criteria and by PCR amplification and sequencing of the β-tubulin gene as previously described.1,2 Antifungal susceptibility testing was performed using the broth microdilution method according to the CLSI M38A2 document for itraconazole (ITC), voriconazole (VRC), posaconazole (PSC), amphotericin B (AMB) and caspofungin (CSF).3CYP51A gene (encoding the 14-α-sterol demethylase, target of azole drugs) was PCR amplified including 5′ UTR, ORF and 3′ UTR regions. The obtained amplification fragment was sequenced as described before.4,5
The A. fumigatus strain MICs and MEC were ITC >8μg/ml, VRC 0.5μg/ml, PSC 0.12μg/ml, AMB 0.5μg/ml and CSF 0.5μg/ml. According to the ECV values published by Espinel-Ingroff et al., the strain was considered as ITC-resistant and susceptible to all the other antifungal agents tested.6,7CYP51A sequencing showed two nucleotide mutations (G161A and G162A) when compared with the one published under the GenBank sequence AF338659.1. These mutations lead to a substitution at codon 54 (G54E) (Fig. 1) which was already described and associated to ITC-resistant phenotypes, but was never seen in South America before.4,8–11 Thus, to the best of our knowledge, this is the first ITC-resistant A. fumigatus strain reported in this part of the world.
CYP51A DNA sequencing chromatogram and Cyp51Ap amino acid sequence for the ITC-resistant A. fumigatus strains. Upper line: Segment of the wild type A. fumigatus Cyp51Ap (GenBank accession no. AAK73659.1) between the amino acid residues 48 and 65. Lower line: the same Cyp51Ap segment of the ITC-resistant strain, showing the amino acid substitution (G54E). The red box in the DNA sequencing chromatogram shows the mutated codon 54.
Azole-resistant A. fumigatus isolation frequency is increasing. These resistant strains were isolated with or without previous azole exposure.8,9,12–14 Our patient, who had never been treated previously with azole drugs, suffered an infection with an ITC-resistant A. fumigatus strain due to an accidental inoculation with vegetal detritus. This fact suggests an environmental origin of the strain. There have been no reports in Argentina demonstrating the existence of environmental azole-resistant A. fumigatus strains. However, these strains might be being selected since azole antifungal agents are widely used in agriculture for plant protection. Such environmental route of resistance development in A. fumigatus was firstly proposed in 2001 and molecularly confirmed later in the Netherlands for a common azole resistance mechanism involving L98H substitution at Cyp51Ap coupled with a promotor modification.15–17
The emergence and spread of the resistance mechanism described here in A. fumigatus is of major concern because ITC is a highly used azole drug in developing countries. It would be useful to analyze environmental sources to detect these strains.
Conflict of interestNone to declare.
This work was supported in part by PICT/PRH N° 2009-0091 and PICT N° 2013-1571 from FONCyT to GGE. FL and CD are doctoral fellows from CONICET and DM is doctoral fellow from FONCyT.