Cryptococcus gattii is a pathogenic basidiomycetous yeast that is emerging in temperate climate zones worldwide. C. gattii has repetitively been isolated from numerous tree species. Ongoing environmental sampling and molecular characterization is essential to understand the presence of this primary pathogenic microorganism in the Mediterranean environment.
AimsTo report the first isolation of the rare C. gattii genotype AFLP7/VGIV from the environment in Europe.
MethodsSamples were collected from woody debris of carob trees (Ceratonia siliqua) and olive trees (Olea europaea) in El Perelló, Tarragona, Spain. Cryptococcus species were further characterized by using URA5-RFLP, MALDI-TOF, AFLP and MLST. The antifungal susceptibility profile to amphotericin B, 5-fluorocytosine, fluconazole, itraconazole, posaconazole and voriconazole was determined using Sensititre Yeast One and E-test.
ResultsCultures from one carob tree revealed the presence of ten Cryptococcus-like colonies. One colony was identified as C. gattii, and subsequent molecular characterization showed that it was an α mating-type that belonged to the rare genotype AFLP7/VGIV. Antifungal susceptibility testing showed values within the range of sensitivity described for other isolates of the same genotype and within the epidemiological cutoff values for this species.
ConclusionsThe isolation of the rare C. gattii genotype AFLP7/VGIV in Spain is the first report in the European environment, implying the possible presence in other regions of the Mediterranean area, and underlines that clinicians must be aware for C. gattii infections in healthy individuals.
Cryptococcus gattii es una levadura perteneciente a los basidiomicetos y considerada un patógeno emergente en climas templados. C. gattii se ha aislado en un gran número de especies de árboles en todo el mundo. El muestreo medioambiental y la caracterización molecular de C. gattii es esencial para entender la presencia de este patógeno primario en el entorno de la cuenca mediterránea.
ObjetivosComunicar la presencia del raro genotipo AFLP7/VGIV de C. gattii en el medioambiente en Europa.
MétodosSe tomaron muestras de detritus de algarrobos (Ceratonia siliqua) y olivos (Olea europaea) en las afueras de El Perelló (Tarragona, España). La colonia de C. gattii aislada se sometió a análisis mediante URA5-RFLP, MALDI-TOF, AFLP y MLST. Se llevó a cabo un estudio de sensibilidad in vitro a la anfotericina B, 5-fluorocitosina, fluconazol, itraconazol, posaconazol y voriconazol mediante las pruebas comerciales Sensititre Yeast One® y E-test®.
ResultadosDe una de las muestras de un algarrobo se aislaron 10 colonias susceptibles de ser Cryptococcus. Una de ellas fue estudiada e identificada como C. gattii, y su subsecuente caracterización molecular mostró que se trataba de un tipo sexual α y que pertenecía al raro genotipo AFLP7/VGIV. El estudio de la sensibilidad a los antifúngicos mostró valores similares a los de otras cepas del mismo genotipo y dentro del rango de valores de corte epidemiológicos para la especie.
ConclusionesEl aislamiento en España de C. gattii con el genotipo AFLP7/VGIV es el primero descrito en el medioambiente en Europa; podría encontrarse también en otros países de la cuenca mediterránea, donde debería tenerse un especial cuidado por la posibilidad de infección en individuos no inmunodeprimidos.
Cryptococcus gattii is a pathogenic basidiomycetous yeast that mainly causes infection by the inhalation of the desiccated yeast cells or basidiospores, in which small size allows them to pass the lung alveoli.25 During the past two decades the number of C. gattii infections has increased in regions that are within temperate climate zones, mainly due to several large ongoing outbreaks in North America.11,12,24 Since the onset of the HIV/AIDS-pandemic during the early 1980s, cryptococcal infections among immunocompromised individuals have raised dramatically. It has been estimated that annually approximately one million HIV-infected patients develop cryptococcal meningitis, and that approximately 625,000 people die due to this fungal infection.23
Cryptococcus neoformans and C. gattii differ in their host predilection, ecology and physiology.2,12,24 The former has a global distribution while C. gattii was restricted to tropical and sub-tropical climate zones,1,11,12,24 but C. gattii is nowadays emerging in temperate climate zones similar to that of Mediterranean Europe.11 Until recently it was believed that certain Eucalyptus species were the exclusive ecological niche for C. gattii, but this assumption was refuted by large-scale environmental screening initiated after several C. gattii outbreaks.4,7,15,24
Within the C. neoformans/C. gattii species complex 13 genotypes can be discerned by using molecular techniques such as PCR fingerprinting, restriction fragment length polymorphism (RFLP) fingerprinting (using the PLB1 and URA5 loci), amplified fragment length polymorphism (AFLP) fingerprinting, multi-locus microsatellite (MLMT) and multi-locus sequence typing (MLST).2,12,13,18C. neoformans can be classified into five genotypes: AFLP1/VNI, AFLP1A/VNII/VNB and AFLP1B/VNII for C. neoformans variety grubii (serotype A), AFLP2/VNIV for C. neoformans variety neoformans (serotype D) and AFLP3/VNIII for the hybrid form (serotype AD).2,18C. gattii can be split into five genotypes known as AFLP4/VGI, AFLP6/VGII and AFLP10/VGIV for serotype B isolates, and AFLP5/VGIII and AFLP7/VGIV for serotype C isolates.12,13,18 Isolates with genotypes AFLP5/VGIII, AFLP7/VGIV and AFLP10/VGIV have frequently been isolated from immunocompromised individuals; on the contrary, the other C. gattii genotypes are often isolated from individuals that apparently do not have any underlying disease.3,12,13,16,24 Although rarely found, interspecies hybrids between haploid C. gattii and C. neoformans genotypes exist, and so far three interspecies hybrid genotypes (all originated from clinical sources) have been described.14
In Europe, infections with C. gattii are being increasingly reported, with the Mediterranean area being the hotspot for autochthonous acquired infections, while among northern European citizens infections were found to be acquired elsewhere.7,12 Animals have been reported as being sentinels for the environmental presence of C. gattii,1,7,20,21 and several outbreaks of C. gattii infections among animals have been reported from the Iberian Peninsula.
In July 2013 environmental samples were taken, as described before,7 as part of ongoing efforts to investigate the presence of C. gattii in the Spanish environment from four carob trees (Ceratonia siliqua) and two olive trees (Olea europaea) located in the outskirts of El Perelló, Tarragona, Spain (40°52′04.5″N 0°43′00.9″E).
The sample of one carob tree (two samples were taken) yielded yeast colonies in the laboratory. Standard phenotypic identification, including canavanine-glycine-bromothymol blue medium, showed that one isolate (CCA436) was C. gattii while the others were unable to grow at 37°C and, therefore, were not considered potentially pathogenic yeasts. Identification was confirmed by MALDI-TOF analysis (Bruker Daltonics, Bremen, Germany) that gave a hit with C. gattii. Mating-type analysis was performed as previously described12 and was positive for the α mating-type. The genotype was determined by RFLP fingerprinting of the URA5-locus and confirmed by AFLP fingerprinting as previously described,7,12,13 that revealed the C. gattii isolate belonged to the rare genotype AFLP7/VGIV.
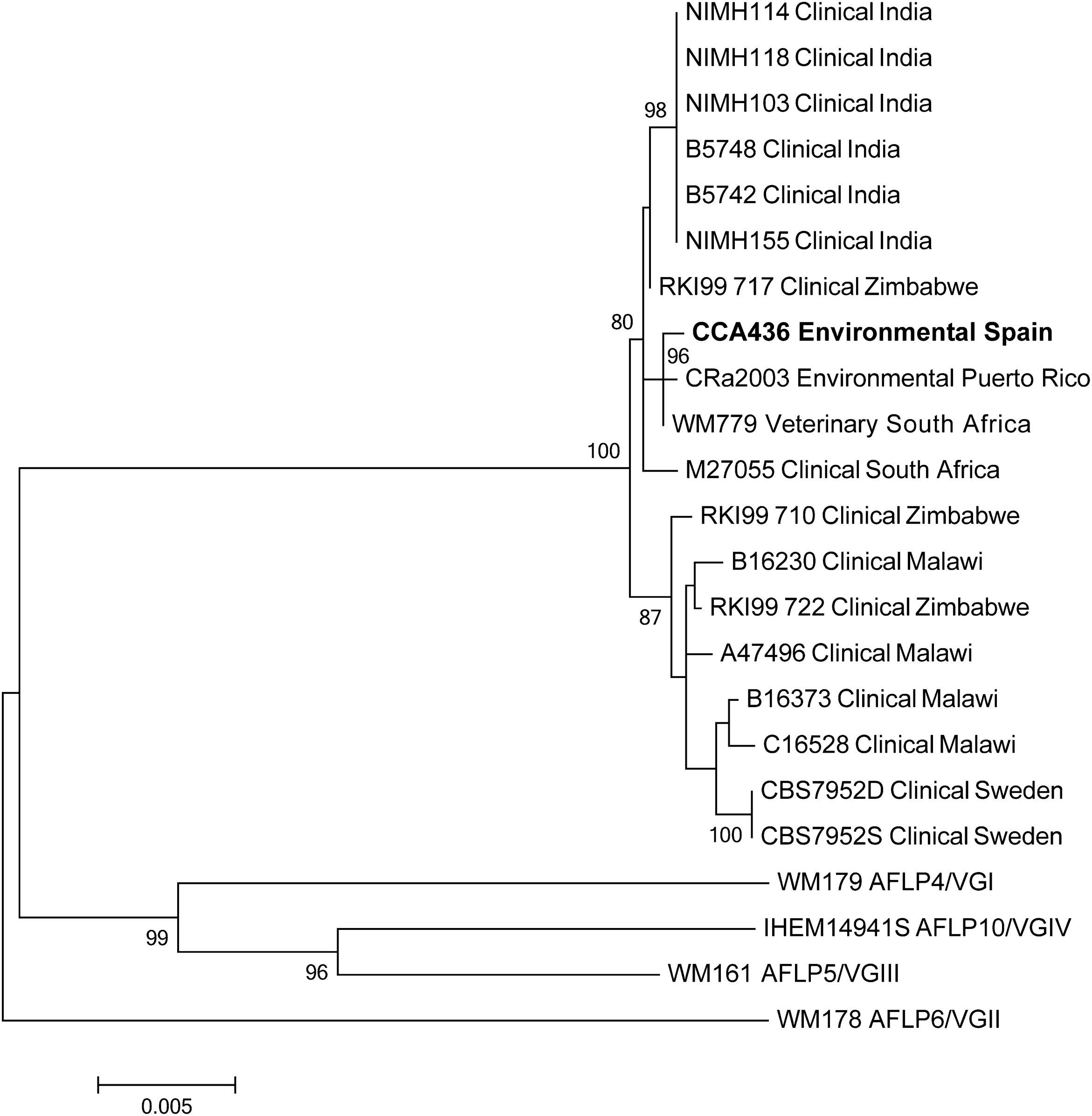
MLST was carried out as described before,12 the CAP59, GPD1, IGS1, LAC1, PLB1, SOD1 and URA5 loci were partly amplified and bi-directionally sequenced for the Spanish isolate CCA436 and for the Caribbean isolate CRa2003.17 Sequences can be accessed via Genbank KM230244, KM230245, KM230255, KM230256, KM230266, KM230267, KM230277, KM230278, KM230288, KM230289, KM230299, KM230300, KM230310 and KM230311. Phylogenetic analysis was performed as described before,12 and published MLST-data obtained from previous C. gattii AFLP7/VGIV reports were included.5,12,17 Phylogenetic analysis showed that isolate CCA436 was closely related to WM799, which came from a South African captive cheetah, and to the Puerto Rican environmental isolate CRa2003 (Fig. 1).
Bootstrapped maximum likelihood phylogenetic tree of known C. gattii AFLP7/VGIV isolates based on multi-locus sequence typing of the 7 loci CAP59, GPD1, IGS1, LAC1, PLB1, SOD1 and URA5. Robustness of the branches is indicated for bootstrap-values of ≥75. The Spanish isolate is indicated in bold.
In vitro antifungal susceptibility testing was performed using the SensiTitre Yeast One method (TREKDS, East Grinstead, United Kingdom), and resulted in the following susceptibility values: amphotericin B 0.5μg/ml, 5-fluorocytosine 2μg/ml, fluconazole 8μg/ml, itraconazole 0.06μg/ml, posaconazole 0.125μg/ml, and voriconazole 0.06μg/ml. E-test (Liofilchem, Roseto degli Abruzzi, Italy) revealed the following values: amphotericin B 1μg/ml, fluconazole 32μg/ml, posaconazole 0.25μg/ml, voriconazole 0.125μg/ml, and 5-fluorocytosine (bioMérieux, Madrid, Spain) >32μg/ml. These values are within the range of sensitivity described for other isolates of the same genotype13 and within the epidemiological cutoff values (ECVs) proposed for the C. gattii/C. neoformans species complex.8–10
The C. gattii isolate has been deposited in the yeast culture collection of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, under accession number CBS12983.
In the current study we report the environmental isolation of a rare C. gattii mating-type α, genotype AFLP7/VGIV isolate from a carob tree, which has only once before been reported from an environmental source in Puerto Rico.17 Clinical C. gattii genotype AFLP7/VGIV cases have been reported from Africa,16,22 India,5 and a travel-related case from Sweden.12 Based on PCR fingerprinting this genotype has been observed as cause of cryptococcal disease in Latin America.19 Subsequent molecular characterization revealed that these clinical Latin American isolates formed a polyphyletic cluster22 and support recent data that the PCR fingerprint molecular type VGIV can be separated into two distinct clusters by AFLP and MLST genotyping, namely AFLP7/VGIV for serotype C isolates and AFLP10/VGIV for serotype B isolates.12,13
From the current report, as well as from others, it is clear that genotype AFLP7/VGI and its sibling AFLP10/VGIV are the least reported and least studied genotypes within the C. gattii/C. neoformans species complex. The occurrence of these rare genotypes remains enigmatic, but ongoing local environmental sampling initiatives, such as the current study and the recently initiated European Environmental C. gattii Sampling Initiative,6 will reveal more insights into the life-style of these rarely reported C. gattii genotypes.
Conflict of interestThe authors declare that there is no conflict of interest.
The authors would like to acknowledge Fundación Navarro-Tripodi (Alicante, Spain) for the financial support and Vishnu Chaturvedi and Sudha Chaturvedi for providing genomic DNA of the Puerto Rican environmental C. gattii genotype AFLP7/VGIV isolate.