Pseudomonas aeruginosa is an important human pathogen that causes severe infections in a wide range of immunosuppressed patients. Herein, we evaluated the proteolytic profiles of 96 Brazilian clinical isolates of P. aeruginosa recovered from diverse anatomical sites.
MethodsCell-associated and extracellular proteases were evidenced by gelatin–SDS–PAGE and by the cleavage of soluble gelatin. Elastase was measured by using the peptide substrate N-succinyl-Ala-Ala-Ala-p-nitroanilide. The prevalence of elastase genes (lasA and lasB) was evaluated by PCR.
ResultsBacterial extracts were initially applied on gelatin–SDS–PAGE and the results revealed four distinct zymographic profiles as follows: profile I (composed by bands of 145, 118 and 50kDa), profile II (118 and 50kDa), profile III (145kDa) and profile IV (118kDa). All the proteolytic enzymes were inhibited by EDTA, identifying them as metalloproteases. The profile I was the most detected in both cellular (79.2%) and extracellular (84.4%) extracts. Overall, gelatinase and elastase activities measured in the spent culture media were significantly higher (around 2-fold) compared to the cellular extracts and the production level varied according to the site of bacterial isolation. For instance, tracheal secretion isolates produced elevated amount of gelatinase and elastase measured in both cellular and extracellular extracts. The prevalence of elastase genes revealed that 100% isolates were lasB-positive and 85.42% lasA-positive. Some positive/negative correlations were showed concerning the production of gelatinase, elastase, isolation site and antimicrobial susceptibility.
ConclusionThe protease production was highly heterogeneous in Brazilian clinical isolates of P. aeruginosa, which corroborates the genomic/metabolic versatility of this pathogen.
Pseudomonas aeruginosa (P. aeruginosa) es un importante patógeno humano que causa graves infecciones en diversos tipos de pacientes inmunodeprimidos. En este trabajo evaluamos los perfiles proteolíticos de 96 aislamientos clínicos brasileños de P. aeruginosa aislados de diferentes localizaciones anatómicas.
MétodosLas proteasas extracelulares y de extractos celulares fueron analizadas por SDS-PAGE copolimerizada con gelatina y a través de clivaje de gelatina en solución. La elastasa fue medida usando el substrato peptídico N-succinil-Ala-Ala-Ala-p-nitroanilida. La prevalencia de genes codificantes para elastasa (lasA y lasB) fue evaluada por PCR.
ResultadosEn primer lugar, los extractos de las bacterias fueron aplicados en geles de SDS-PAGE-gelatina, los cuales, después de revelados, revelaron 4 perfiles enzimográficos, así: perfil i (compuesto por bandas de 145, 118 y 50kDa), perfil ii (118 y 50kDa), perfil iii (145kDa) y perfil iv (118kDa). Todas las enzimas proteolíticas fueron inhibidas por EDTA, siendo, por tanto, identificadas como metaloproteasas. El perfil i fue el más detectado tanto en los extractos celulares (79,2%) como en los extracelulares (84,4%). Las actividades de gelatinasa y elastasa medidas en el medio de cultivo fueron significativamente más elevadas (cerca de 2 veces) que en los extractos celulares y el nivel de producción varió de acuerdo al sitio del cual fue aislada la cepa. Por ejemplo, cepas aisladas de secreción traqueal produjeron cantidades elevadas de gelatinasa y elastasa medidas tanto en el extracto celular como en los extractos extracelulares. La prevalencia de los genes de elastasa reveló que el 100% de los aislamientos fueron lasB positivos y 85.42% lasA positivos. En algunos casos se observó una correlación positiva/negativa respecto a la producción de gelatinasa, elastasa, sitio de aislamiento y susceptibilidad antimicrobiana.
ConclusiónLa producción de proteasas fue altamente heterogénea en los aislamientos clínicos brasileños de P. aeruginosa, lo cual corroboran la versatilidad genómica/metabólica de este patógeno.
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium able to grow and survive in almost all environments, living primarily in water, soil, vegetation and both human and animal sewages.1,2 Moreover, P. aeruginosa is a human pathogen frequently isolated in worldwide hospital settings, causing numerous debilitating nosocomial infections, especially in immunocompromised individuals and patients with cancer, cystic fibrosis and burns.1,3 The pathogenesis of P. aeruginosa is a complex and multifactorial event, which can be faced as a typical war battle: on one hand, the production of attributes of bacterial virulence and, on the other hand, the host's ability to plan a vigorous counterattack by generating a powerful immune response.4,5
A multitude of virulence factors are produced by P. aeruginosa cells, including: adhesins (e.g., flagella and type 4 pili), endotoxin (e.g., lipopolysaccharide), exotoxins (e.g., exotoxin A), pigments (e.g., pyocyanin), siderophores (e.g., pyoverdine), rhamnolipids, alginate-formed biofilm and a plentiful of extracellular hydrolytic enzymes (e.g., phospholipases, esterases, lipases and proteases).4–6 Collectively, all these bacterial virulence attributes act by increasing the tissue damage and/or protecting this pathogen against the host immune system recognition, contributing to the establishment and maintenance of the infectious process.4–6 Among all the virulence factors described in P. aeruginosa, proteases seem to play central physiological roles and particularly important functions in different stages of the bacteria-host interplay. Corroborating this statement, of the 5568 open reading frames encoded in the genome of the P. aeruginosa type strain PAO1, 155 (2.8%) are listed as proteases and, in addition, the manifold secretory systems synthesized by this bacterial pathogen churn out vast quantities of proteases.7 The foremost well-characterized proteases produced by P. aeruginosa are: elastases A (Las A) and B (Las B), alkaline protease and protease IV.8,9Pseudomonas elastases and alkaline protease are metallo-type proteases that can degrade a variety of proteins, including a huge range of extracellular matrix components, defense molecules and proteinaceous surfactants encountered in several cells, tissues and fluids of the host.9,10 Also, this pathogen secretes protease IV, a serine-type protease, widely found in P. aeruginosa clinical isolates from ocular infections.11,12
Despite the importance of proteases in both physiological and pathological events of P. aeruginosa cells, very little is known on the production/expression of these host-damaging enzymes taking into consideration the anatomical site of infection. In this context, the aim of the present work was to evaluate the cellular and extracellular proteases produced by 96 clinical isolates of P. aeruginosa, which were recovered from distinct anatomical sites of patients attended at Brazilian hospitals.
MethodsClinical strainsThe present study was conducted with 96 non-duplicated clinical strains of Pseudomonas aeruginosa isolated from rectum (n=20), tracheal aspirate (n=19), mouth (n=18), blood (n=8), urine (n=7), central venous catheter (n=6), pleural secretion (n=5), eschar (n=4), cystic fibrosis lungs (n=4), sputum (n=3) and nasal secretion (n=2) of patients hospitalized in intensive treatment units of four hospitals located in the Southeast States of Brazil.13–15 Each P. aeruginosa isolate used herein came from only one patient; consequently, each isolate was recovered from only one anatomical site regarding each patient. Regarding the isolation site, we separated our sample collection in two major groups: (i) isolates from sites (urine, eschar, blood, pleural secretion, sputum, venous catheter tip, nasal secretion and cystic fibrosis lung) linked with bacterial infection, in which the patients presented signs and symptoms (e.g., fever, pus from wound, pneumonia, etc.) and (ii) isolates from sites (tracheal secretion, mouth and rectum) associated to bacterial colonization, in which the patients had no signs and/or symptoms.15 The genetic variability and antimicrobial susceptibility profiles of the P. aeruginosa isolates used in all parts of the current study were recently published by our research group.15 The reference strain of P. aeruginosa ATCC 27853 was used as a control in all experiments.
Growth conditions and bacterial extractsP. aeruginosa strains were grown on trypticase soy agar (TSA; Merck, Darmstadt, Germany) for 18h at 37°C. Subsequently, bacterial cells were subcultured in tryptone soy broth (TSB) supplemented with 1% glycerol, 50mM glutamate, 10mM CaCl2 and 10mM ZnCl2 and incubated for 24h at 37°C under constant agitation.9 Cultures were centrifuged (4000rpm, 20min, 4°C) and bacterial cells were washed three times in phosphate-buffered saline (PBS; 150mM NaCl, 20mM phosphate buffer, pH 7.2). Then, bacteria were suspended in 500μl of PBS supplemented with 0.1% Triton X-100 and lysed in a cell homogenizer (Braun Biotech International, Germany) by alternating 2min shaking periods and 2min cooling intervals (total of 5 cycles). The mixtures were centrifuged (4000rpm, 20min, 4°C) and the obtained supernatants were considered as bacterial cellular extracts.16 In parallel, the spent culture supernatants were filtered in a 0.22-μm membrane (Millipore, São Paulo, Brazil). The cell-free culture supernatants were concentrated 10-fold in a 10,000 molecular weight cut-off Centricon micropartition system (Amicon, Beverly, MA, USA). The protein concentration was determined by the method described by Lowry et al. (1951),17 using bovine serum albumin (BSA, Sigma–Aldrich, USA) as standard.
Zymography assayThe protease profile was assayed by means of dodecyl sulfate sodium-polyacrylamide gel electrophoresis (SDS–PAGE) containing 0.1% gelatin incorporated into the gel as proteinaceous substrate.18 Gels were loaded with 40μg of proteins per slot. After electrophoresis at constant current of 120V at 4°C, SDS was removed by incubation with 10 volumes of 2.5% Triton X-100 for 1h at room temperature under constant agitation. In order to promote the proteolysis, the gels were incubated for 24h at 37°C in the digestion buffer containing 50mM Tris–HCl, 10mM CaCl2, 1mM ZnCl2 and 150mM NaCl, pH 8.0,9 in the presence and in the absence of ethylenediaminetetraacetic acid (EDTA) at 10mM. The gels were stained for 2h with 0.2% Coomassie brilliant blue R-250 in methanol:acetic acid:water (50:10:40) and destained overnight in a solution containing methanol:acetic acid:water (5:10:85), to intensify the digestion halos. The molecular masses of the proteases were calculated by comparison with the mobility of low molecular mass standards. The gels were dried, scanned and digitally processed.16
In-solution gelatinase activity assayThe gelatinase activity was measured in both cellular and spent culture supernatant extracts of each P. aeruginosa isolate, according to the method described by Buroker-Kilgore and Wang.19 The bacterial extracts (equivalent to 20μg of protein) were mixed with 70μl of 1% gelatin and 260μl reaction buffer (50mM Tris–HCl, 10mM CaCl2, 1mM ZnCl2, 150mM NaCl, pH 8.0). The reaction mixtures were incubated for 2h at 37°C. After that, three aliquots (100μl each) of the reaction mixture were transferred to wells of a microtiter plate containing 50μl of water and 100μl of a Coomassie solution (0.025% Coomassie brilliant blue G-250, 11.75% ethanol, and 21.25% phosphoric acid). After 10min to allow dye binding, the plate was read on a Thermomax Molecular Device microplate reader at an absorbance of 595nm. One unit of gelatinase activity was defined as the amount of enzyme that caused an increase of 0.01 in the absorbance unit.20
In-solution elastase assayElastase activity was assayed using a microtiter-based assay, which consisted on the cleavage of an elastase-specific chromogenic peptide substrate, N-succinyl-Ala-Ala-Ala-p-nitroanilide (Sigma–Aldrich, USA) as described by Kocabiyik et al. (1995).21 The reaction mixtures (100μl) containing each cellular extract or cell-free culture supernatant (20μg of proteins), chromogenic substrate at 1mM and buffer (50mM Tris–HCl, 10mM CaCl2, 1mM ZnCl2, 150mM NaCl, pH 8.0) were incubated for 2h at 37°C in a 96-well microplate. Then, the plate was read on a Thermomax Molecular Device microplate reader at an absorbance of 405nm. One unit of elastase activity was defined as the amount of enzyme that caused an increase of 0.01 in the absorbance unit.20
Detection of elastase genesIn order to extract the bacterial DNA, the P. aeruginosa clinical isolates were grown on TSA for 18h at 37°C. Afterward, three colonies were suspended in 1ml of 0.1M PBS and centrifuged at 12,000rpm for 5min. Supernatants were discarded and the pellets were resuspended in 100μl of ultrapure water, boiled for 8min and then remained on ice for 30min. The mixtures were centrifuged at 12,000rpm for 5min and the pellets were discarded, while the supernatants containing the DNA were stored at −20°C22 prior to polymerase chain reaction (PCR) for the detection of the genes encoding elastase A (lasA) and elastase B (lasB) of P. aeruginosa. In this set of experiments, the primer sequences used were previously described by Lanotte et al. (2004).23 Gene amplification was performed in a 25-μl mixture containing 12.5μl of Green Go Taq master mix (0.05U/μl Taq DNA polymerase, reaction buffer, 4mM MgCl2, 0.4mM of each dNTP) (Promega, USA), 1μl of lasA primer (forward, 5′-CGCCATCCAACCTGATGCAAT-3′ and reverse, 5′-AGGCCGGGGTTGTACAACGGA-3′) and lasB primer (forward: 5′-GGAATGAACGAAGCGTTCTC-3′ and reverse, 5′-GGTCCAGTAGTAGCGGTTGG-3′)23 at 10pmol/μl (Exxtend, Brazil), and 1μl of DNA template. The PCR conditions were as follow: initial denaturation at 94°C for 10min; 30 cycles of 94°C for 1min, 60°C (lasA) or 55°C (lasB) for 1min, 72°C for 1min and 72°C for 5min using Arktik Thermal cycler (Thermo Scientific, USA). The PCR products were analyzed by electrophoresis with 1% agarose gels in 0.5× Tris-borate-EDTA buffer (TBE; 89mM Tris, 89mM boric acid, 2mM EDTA, pH 8.0) running buffer at 100V for 1h. The gels were stained with 0.5mg/ml of ethidium bromide and the amplicons corresponding to lasA (600 pb) and lasB (300 pb) genes were detected under UV transillumination (Loccus Biotecnoligia, São Paulo, Brazil).
Statistics and correlationsAll the experiments were performed in triplicate, in three independent experimental sets. The data were expressed as mean±standard deviation. The results were evaluated by analysis of variance (ANOVA) using Graphpad Prism 5 computer software. The intergroup comparison (resistant versus susceptible strains) was performed by using Fisher's exact test using SPSS Statistics program. The correlation tests were determined by Pearson's correlation coefficient (r). In all analyses, P values of 0.05 or less were considered statistically significant.
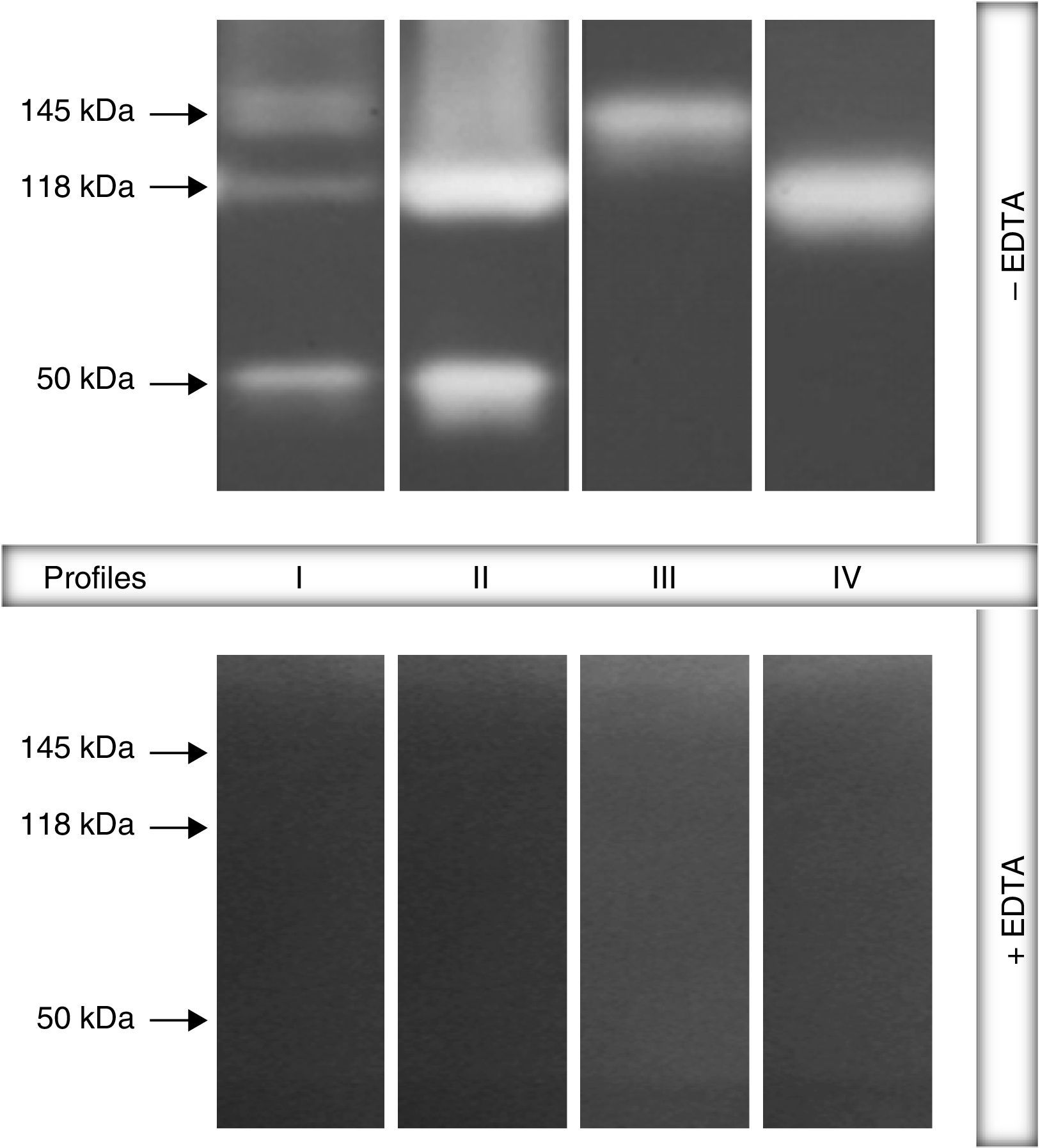
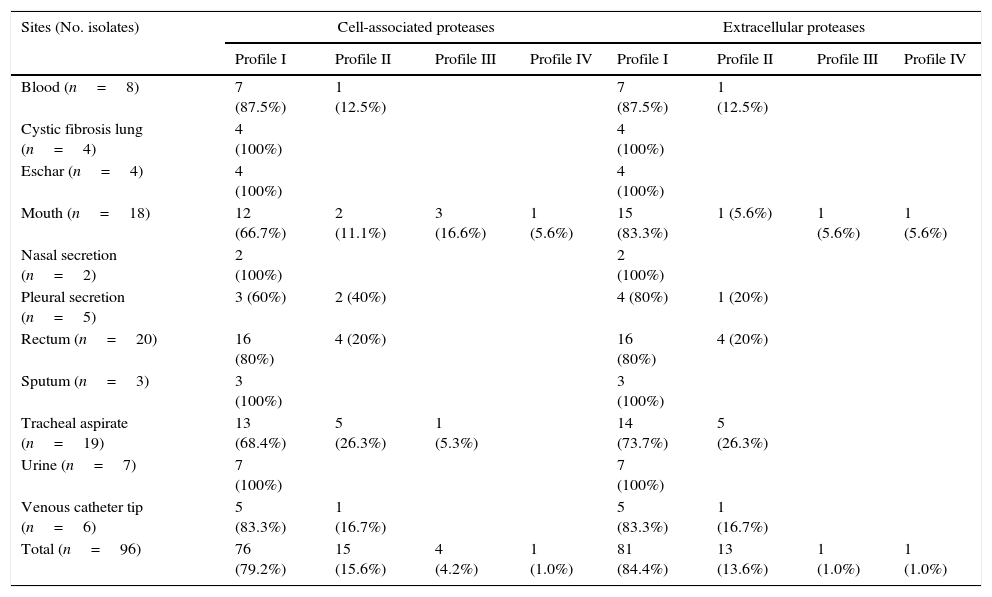
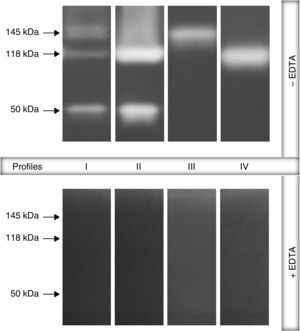
ResultsGelatin–SDS–PAGE profilesAll the clinical isolates of P. aeruginosa were able to produce both cell-associated and extracellular proteases, as judged by the gelatin hydrolysis detected in co-polymerized gels; however, distinct profiles were clearly observed. In this sense, four zymographic profiles, composed by proteolytic bands with molecular masses ranging from 50 to 145kDa, were evidenced as following: profile I, formed by proteolytic bands of 145, 118 and 50kDa; profile II, proteolytic bands of 118 and 50kDa; profile III, a single band of 145kDa; profile IV, a single band of 118kDa (Fig. 1). Moreover, EDTA at 10mM was able to completely inhibit all the proteolytic activities evidenced in the zymogram gels, classifying them as metallo-type proteases (Fig. 1). Interestingly, the profile I was the most predominant protease profile observed in the vast majority of both cellular (76/96; 79.2%) and extracellular (81/96; 84.4%) extracts obtained from P. aeruginosa isolates (Table 1), being the unique profile detected in clinical isolates from urine, eschar, sputum, nasal secretion and lung of cystic fibrosis patients (Table 1).
Representative proteolytic profiles observed in Brazilian clinical isolates of Pseudomonas aeruginosa from different anatomical sites. The proteases were evidenced by electrophoresis on 10% SDS–PAGE containing 0.1% gelatin as the copolymerized protein substrate as described in “Material and methods” section. Molecular masses of the proteases, expressed in kilodaltons (kDa), are represented on the left. Four different profiles were clearly seen: profile I, bands of 145kDa+118kDa+50kDa; profile II, 118kDa+50kDa; profile III, 145kDa and profile IV, 118kDa. Galdino et al., 2016.
Proteolytic profiles observed in cellular extracts and culture supernatants from Brazilian clinical isolates of Pseudomonas aeruginosa from different isolation sites.
| Sites (No. isolates) | Cell-associated proteases | Extracellular proteases | ||||||
|---|---|---|---|---|---|---|---|---|
| Profile I | Profile II | Profile III | Profile IV | Profile I | Profile II | Profile III | Profile IV | |
| Blood (n=8) | 7 (87.5%) | 1 (12.5%) | 7 (87.5%) | 1 (12.5%) | ||||
| Cystic fibrosis lung (n=4) | 4 (100%) | 4 (100%) | ||||||
| Eschar (n=4) | 4 (100%) | 4 (100%) | ||||||
| Mouth (n=18) | 12 (66.7%) | 2 (11.1%) | 3 (16.6%) | 1 (5.6%) | 15 (83.3%) | 1 (5.6%) | 1 (5.6%) | 1 (5.6%) |
| Nasal secretion (n=2) | 2 (100%) | 2 (100%) | ||||||
| Pleural secretion (n=5) | 3 (60%) | 2 (40%) | 4 (80%) | 1 (20%) | ||||
| Rectum (n=20) | 16 (80%) | 4 (20%) | 16 (80%) | 4 (20%) | ||||
| Sputum (n=3) | 3 (100%) | 3 (100%) | ||||||
| Tracheal aspirate (n=19) | 13 (68.4%) | 5 (26.3%) | 1 (5.3%) | 14 (73.7%) | 5 (26.3%) | |||
| Urine (n=7) | 7 (100%) | 7 (100%) | ||||||
| Venous catheter tip (n=6) | 5 (83.3%) | 1 (16.7%) | 5 (83.3%) | 1 (16.7%) | ||||
| Total (n=96) | 76 (79.2%) | 15 (15.6%) | 4 (4.2%) | 1 (1.0%) | 81 (84.4%) | 13 (13.6%) | 1 (1.0%) | 1 (1.0%) |
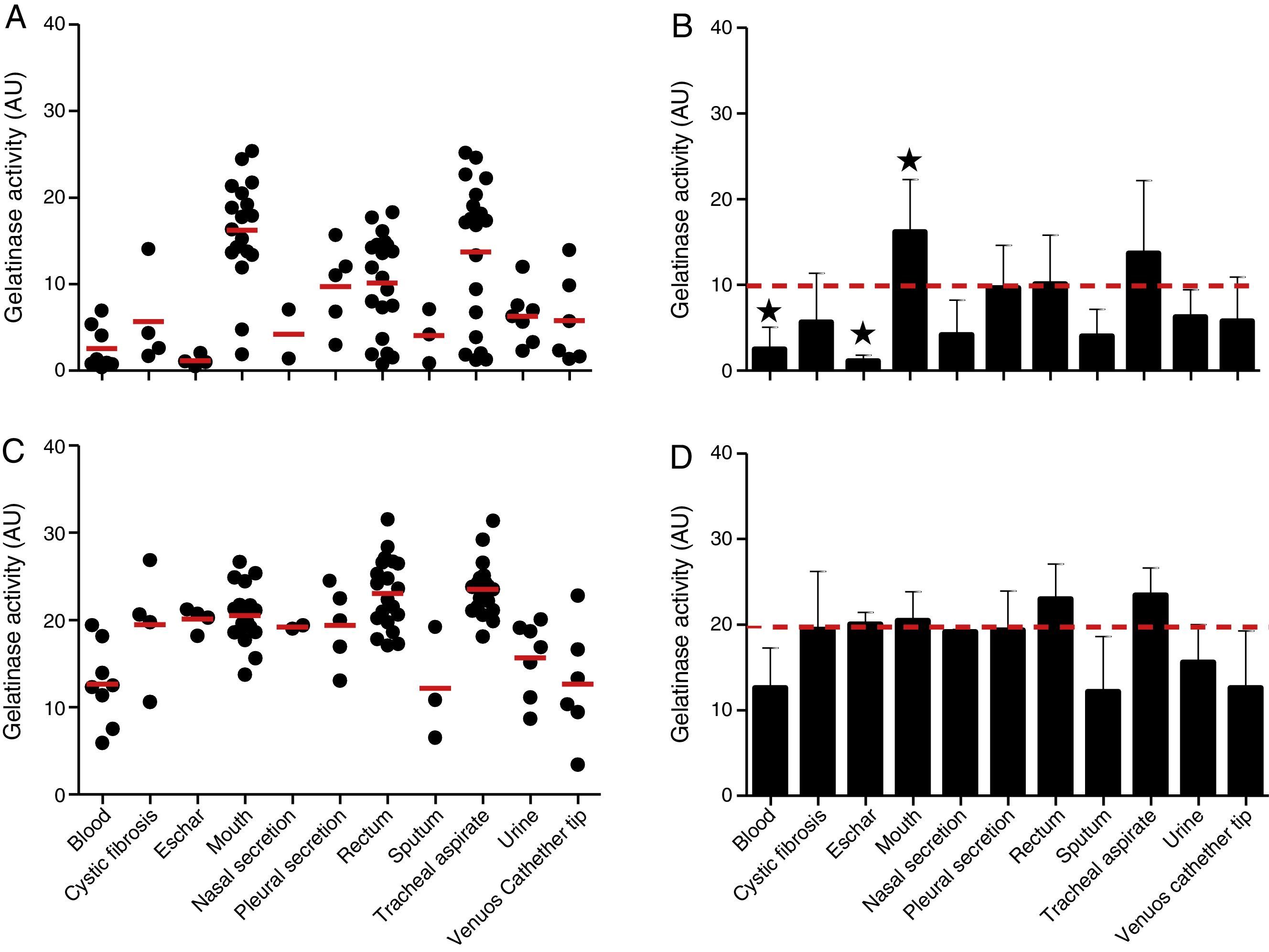
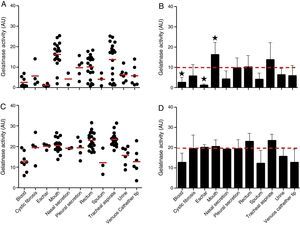
The proteolytic activities in both cellular and extracellular extracts from P. aeruginosa were quantified by using soluble gelatin as protein substrate (Fig. 2). By this protocol, the mean production of gelatinase activity quantified in the cellular extracts of P. aeruginosa (n=96 strains) was 9.89±7.38 arbitrary units (AU) (ranging from 0.37 to 25.37AU) (Fig. 2A and B). The isolates from mouth showed the highest production of cell-associated gelatinase activity (mean of 16.22±6.07AU) followed by isolates from tracheal secretion (mean of 13.71±8.48AU), while isolates from eschar had the lowest production of gelatinase activity (mean of 1.15±0.66AU) followed by isolates from blood (mean of 2.55±0.89AU) (Fig. 2A and B). In general, the gelatinase activity quantified in the supernatant fluids was significantly higher (2-fold) in comparison to the cell-associated proteolytic activity, presenting mean activity of 19.72±5.61AU (ranging from 3.40 to 31.35AU) (Fig. 2C and D). Isolates from tracheal secretion (mean of 23.51±3.25AU) and rectum (mean of 23.05±4.06AU) presented the highest levels of extracellular gelatinase activity, whereas the isolates from sputum (mean of 12.08±6.45AU), blood (mean of 12.64±4.65AU) and central venous catheter tip (mean of 12.65±6.65AU) had the lowest amounts of extracellular gelatinase activity (Fig. 2C and D).
Gelatinase activity in Brazilian clinical isolates of Pseudomonas aeruginosa from diverse anatomical sites. The proteolytic activity was measured in the cellular and extracellular bacterial extracts using soluble gelatin as the substrate. Distribution of cellular (A) and extracellular (C) gelatinase activity in each clinical isolate according to anatomical isolation site. Mean±standard deviation regarding the cellular (B) and extracellular (D) gelatinase activity in each anatomical site. The red lines in (A, C) indicate the arithmetic average of the production of gelatinase in each anatomical site, while the red dotted line in (B, D) is the overall average for the production of gelatinase considering all studied P. aeruginosa isolates (n=96). Isolates from tracheal secretion and mouth (highest activities) and eschar (lowest activity) showed statistical difference when compared to the overall average considering the 96 clinical isolates (★, P<0.05, one-way ANOVA, Turkey's multiple comparison test). Galdino et al., 2016.
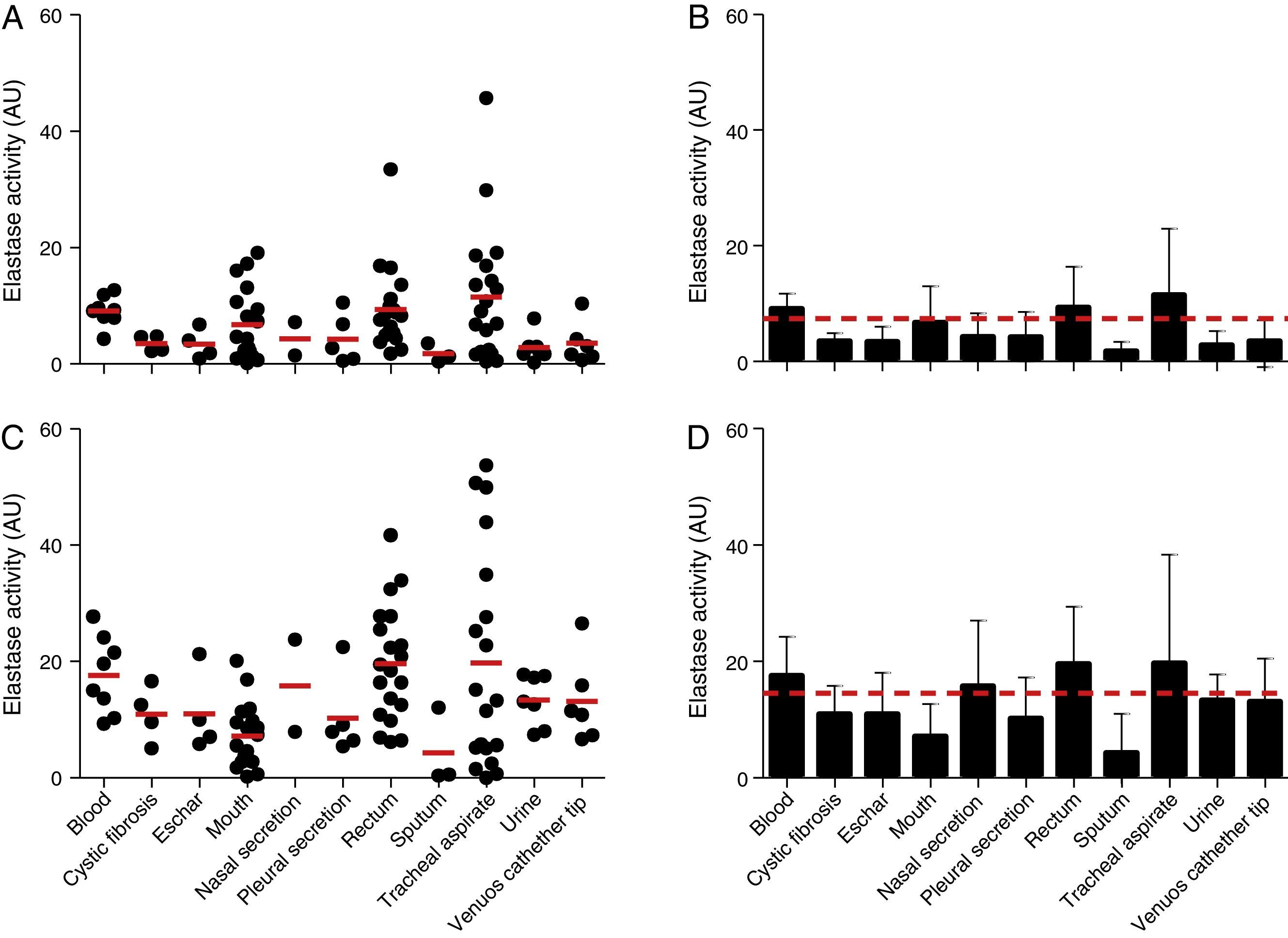
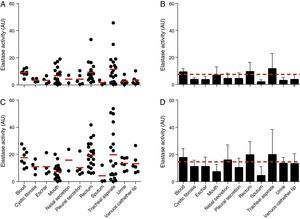
The elastase activity of P. aeruginosa was evaluated using the chromogenic peptide substrate N-succinyl-Ala-Ala-Ala-p-nitroanilide as previously proposed21 (Fig. 3). All the 96 P. aeruginosa clinical isolates were able to produce intracellular and extracellular elastase. Collectively, the cell-associated elastase activity ranged from 0.10 to 45.70AU, with mean of 7.36±7.40AU. Isolates from tracheal secretion showed the highest production of cell-associated elastase activity (mean of 11.52±11.24AU), while the isolates from sputum showed the lowest one (mean of 1.75±1.62AU) (Fig. 3A and B). In addition, all the P. aeruginosa isolates showed extracellular elastase activity with mean production of 14.51±11.49AU (ranging from 0.04 to 53.70AU) (Fig. 3C and D), which was 2-fold higher than the average calculated to the cellular extracts. We observed that isolates from rectum showed the highest extracellular activity (mean of 19.60±9.84AU) followed by isolates from tracheal secretion (mean of 19.38±18.66AU); conversely, isolates from sputum showed the lowest amount of extracellular elastase activity (mean of 4.32±6.69AU) (Fig. 3C and D).
Elastase activity in Brazilian clinical isolates of Pseudomonas aeruginosa from diverse anatomical sites. The proteolytic activity was measured in the cellular and extracellular bacterial extracts using the chromogenic peptide substrate N-succinyl-Ala-Ala-Ala-p-nitroanilide. Distribution of cellular (A) and extracellular (C) elastase activity in each clinical isolate according to anatomical isolation site. Mean±standard deviation regarding the cellular (B) and extracellular (D) elastase activity in each anatomical site. The red lines in (A, C) indicate the arithmetic average of the production of elastase in each anatomical site, while the red dotted line in (B, D) is the overall average for the production of elastase considering all studied P. aeruginosa isolates (n=96). Galdino et al., 2016.
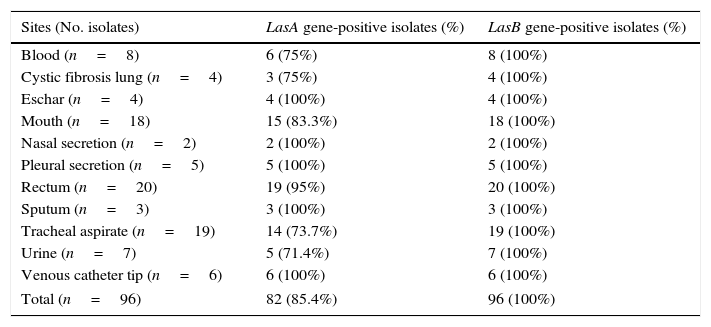
In this set of experiments, the P. aeruginosa isolates were screened for the presence of elastase genes, lasA and lasB, by PCR assay. The lasB gene was successfully amplified in all the studied isolates, while lasA gene was evidenced in 82 (85.42%) of them (Table 2).
Prevalence of LasA and LasB genes in Brazilian clinical isolates of Pseudomonas aeruginosa.
| Sites (No. isolates) | LasA gene-positive isolates (%) | LasB gene-positive isolates (%) |
|---|---|---|
| Blood (n=8) | 6 (75%) | 8 (100%) |
| Cystic fibrosis lung (n=4) | 3 (75%) | 4 (100%) |
| Eschar (n=4) | 4 (100%) | 4 (100%) |
| Mouth (n=18) | 15 (83.3%) | 18 (100%) |
| Nasal secretion (n=2) | 2 (100%) | 2 (100%) |
| Pleural secretion (n=5) | 5 (100%) | 5 (100%) |
| Rectum (n=20) | 19 (95%) | 20 (100%) |
| Sputum (n=3) | 3 (100%) | 3 (100%) |
| Tracheal aspirate (n=19) | 14 (73.7%) | 19 (100%) |
| Urine (n=7) | 5 (71.4%) | 7 (100%) |
| Venous catheter tip (n=6) | 6 (100%) | 6 (100%) |
| Total (n=96) | 82 (85.4%) | 96 (100%) |
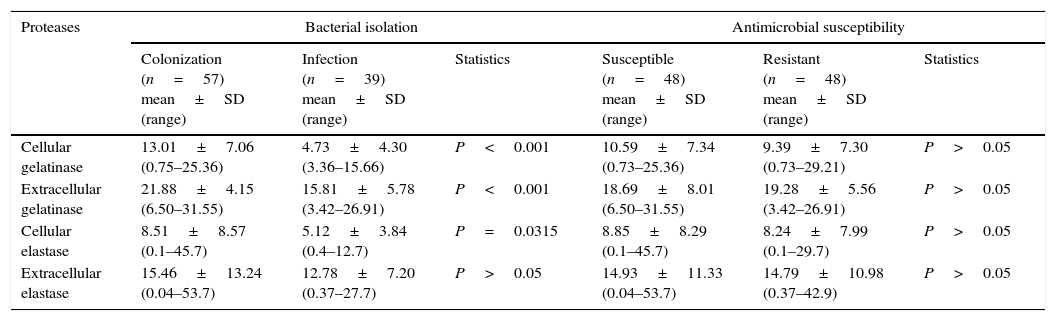
Once the production of gelatinase and elastase from 96 clinical isolates of P. aeruginosa was established, a possible correlation between them was evaluated. In this context, a significant positive correlation was observed to exist between cell-associated and extracellular gelatinase activities (r=0.3728, P=0.0002) (Table 3). On the other hand, when the correlation analysis was performed taking into account the anatomical isolation site of P. aeruginosa, a negative significant correlation was found between cell-associated and extracellular gelatinase activities in isolates from eschar (r=−0.9716, P=0.0284). Additionally, we compared the protease production in P. aeruginosa isolates from sites associated to colonization (e.g., tracheal secretion, mouth and rectum) and isolates from sites correlated with infection (e.g., urine, eschar, blood, pleural secretion, sputum, central venous catheter tip, nasal secretion and cystic fibrosis lung). Considering these parameters, the P. aeruginosa isolates from colonization sites have presented a significantly higher production of both cell-associated and extracellular gelatinase activities (P<0.001) and of cell-associated elastase (P=0.0315) when compared with isolates from infection sites (Table 3). Finally, we have performed a correlation between protease production and antimicrobial susceptibility profile. Among the 96 analyzed isolates, 48 (50%) were susceptible to ceftazidime, imipenem and meropenem, while the other 48 isolates were resistant to these tested antibiotics as previously reported by our research group.15 No significant difference was observed between cellular and extracellular protease production (considering either gelatinase or elastase) regarding the resistant and susceptible P. aeruginosa strains (Table 3).
Correlations among production of gelatinase, elastase, site of bacterial isolation and antimicrobial susceptibility in Brazilian clinical isolates of Pseudomonas aeruginosa.
| Proteases | Bacterial isolation | Antimicrobial susceptibility | ||||
|---|---|---|---|---|---|---|
| Colonization (n=57) mean±SD (range) | Infection (n=39) mean±SD (range) | Statistics | Susceptible (n=48) mean±SD (range) | Resistant (n=48) mean±SD (range) | Statistics | |
| Cellular gelatinase | 13.01±7.06 (0.75–25.36) | 4.73±4.30 (3.36–15.66) | P<0.001 | 10.59±7.34 (0.73–25.36) | 9.39±7.30 (0.73–29.21) | P>0.05 |
| Extracellular gelatinase | 21.88±4.15 (6.50–31.55) | 15.81±5.78 (3.42–26.91) | P<0.001 | 18.69±8.01 (6.50–31.55) | 19.28±5.56 (3.42–26.91) | P>0.05 |
| Cellular elastase | 8.51±8.57 (0.1–45.7) | 5.12±3.84 (0.4–12.7) | P=0.0315 | 8.85±8.29 (0.1–45.7) | 8.24±7.99 (0.1–29.7) | P>0.05 |
| Extracellular elastase | 15.46±13.24 (0.04–53.7) | 12.78±7.20 (0.37–27.7) | P>0.05 | 14.93±11.33 (0.04–53.7) | 14.79±10.98 (0.37–42.9) | P>0.05 |
Proteolytic enzymes have emerged as central attributes associated with virulence of P. aeruginosa due to their multitude of biological roles.4 In the present study, we reported the proteolytic profile obtained from 96 clinical isolates of P. aeruginosa, which were recovered of different anatomical sites from patients attended at Brazilian hospitals, by means of distinct methodological approaches. Firstly, gelatin-containing gels revealed that all the studied bacterial strains produced both cell-associated and extracellular proteases. In this way, four distinct zymogram profiles were clearly identified, being composed by hydrolytic bands with molecular masses of 145, 118 and 50kDa. According to several published papers, elastase B (LasB), elastase A (LasA or staphylolysin) and alkaline protease are represented in gelatin zymograms by proteolytic bands at 145, 118 and 50kDa, respectively.24–26 Furthermore, the results showed that the profile I (zymogram containing the three hydrolytic bands of 145, 118 and 50kDa) was the most frequently found (∼80%) in our P. aeruginosa sample collection. Marquart and co-workers (2005)9 demonstrated that the reference strains of P. aeruginosa (PA01 and ATCC 19660) were able to produce different proteases in gelatin zymogram, including: protease IV (band >200kDa), LasA/LasB (bands of approximately 100kDa) and alkaline protease (51kDa). Schmidtchen and co-workers (2001)27 evaluated the protease activity of 26 P. aeruginosa clinical strains derived from chronic leg ulcers isolated from patients admitted at Swiss hospitals. It was observed that 21/26 (∼81%) displayed proteolytic profile with bands of 100 and 50kDa, which respectively correspond to elastase and alkaline protease.27 In another study, Choy and co-workers (2008)26 analyzed the proteolytic profile of 28 contact lens-related and 27 non-contact lens-related keratitis strains of P. aeruginosa isolated from patients attended at Australian hospitals. Those authors found two different proteolytic profiles in isolates from eye infection that changed according to the source of keratitis: the protease profile I (>200, 145 and 51kDa) was mostly detected in non-contact lens isolates, while proteolytic profile II (>200, 98 and 51kDa) was the most observed in contact lens keratitis isolates.26
Numerous studies have shown that the majority of environmental and clinical isolates of P. aeruginosa exhibited proteolytic activity, particularly elastase activity.27–29 In this context, Moneto Ledesma and co-workers (1985)30 reported that all clinical isolates of P. aeruginosa expressed gelatinolytic activity; however, only 80% of them expressed elastinolytic activity. In other study, Tingpej and co-workers (2007)28 observed that 32 of the 43 (74%) Australian isolates exhibited elastase activity using the elastase Congo red assay, ranging from 3 mU/ml to 367mU/ml. The analysis of virulence factors performed by Jacomé and co-workers (2007)31 revealed that 72.1% (44/61) Brazilian isolates of P. aeruginosa were gelatinase producers. Similarly, our results showed that all the 96 P. aeruginosa strains studied herein were able to degrade gelatin incorporated into the gel matrix or in-solution, considering the evaluation of both bacterial cellular extracts and culture supernatant fluids. Likewise, elastase activity was detected in all the cellular extracts and extracellular supernatants obtained from our P. aeruginosa collection. However, the amount of both gelatinase and elastase activities varied immensely, showing a typically P. aeruginosa isolate-dependence. In addition, some correlations could be done considering the amount of gelatinase/elastase and the site of bacterial isolation. In this way, tracheal secretion isolates produced the highest amount of gelatinase and elastase activities measured in both cellular and extracellular bacterial extracts. Wysockiand co-workers (2013)32 reported that all isolates from venous leg ulcer presented both gelatinolytic and elastinolytic activities. Hamood and co-workers (1996)33 analyzed the production of extracellular virulence factors and showed that isolates from tracheal secretion and urine produced high levels of elastase activity. Woods and co-workers (1986)34 showed that Canadian isolates from acute lung infections showed the highest production of elastase (0.053±0.021mg/ml) compared with elastase activity of isolates from burns, wounds, cystic fibrosis lung and blood.
Among the proteases produced by P. aeruginosa, elastases are assumed to play a major role during acute P. aeruginosa infection.35–37 Corroborating this statement, high prevalence rates of elastase genes in P. aeruginosa were related by several groups.38–41 Using PCR approach, it was evaluated the prevalence of elastase genes in our P. aeruginosa sample collection, revealing that 100% were lasB-positive and 85% lasA-positive isolates.
The production of Pseudomonas proteases might be crucial to cause tissue damage, to degrade the human innate immune system and, finally, to modulate human adaptive immune mechanisms, favoring the establishment of systemic infection or more localized chronic colonization.42 Some studies showed that bacterial isolates recovered from chronic infections were able to produce less extracellular proteases.35 Our results have shown that regardless of the isolation site, some isolates produced significantly higher amounts of elastase. However, in general, isolates from colonization sites (tracheal secretion, mouth and rectum) produced higher levels of gelatinase and elastase than isolates from infection sites (urine, eschar, blood, central venous catheter tip, nasal secretion and cystic fibrosis lung). Similarly to our results, Jagger and co-workers (1986)43 observed that isolates from colonized cystic fibrosis patients presented higher levels of proteolytic activity when compared with chronically infected cystic fibrosis patients.
We analyzed the production of gelatinase and elastase in antimicrobials resistant and susceptible isolates in order to evaluate probable relationships among protease productions and susceptibility to antimicrobial profiles. Our findings showed no difference in gelatinase and elastase production by susceptible and resistant P. aeruginosa clinical isolates (Table 3). Deptula and Gospodarek (2010)44 have compared the production of several virulence factors in susceptible (n=75) and multidrug resistant clinical isolates (n=75) of P. aeruginosa and, contrarily to our findings, the authors observed that P. aeruginosa multidrug resistant isolates exhibited lower elastase activity when compared to susceptible ones. According to Karatuna and co-workers (2010),45 the isolates that were negative for elastase production were generally more resistant to ceftazidime and piperacillin and isolates that exhibited low levels of alkaline protease were more resistant to cefepime, imipenem, tobramycin, piperacillin, piperacillin-tazobactam and ciprofloxacin. Previously, Sánchez and co-workers (2002)46 observed that laboratory-obtained mutants of P. aeruginosa overproducing multidrug efflux pumps (MexABOprM and MexCDOprJ) secreted lower amounts of elastase. These findings indicate that in vivo genetic selection of multidrug resistant strains may lead to changes of genotypic/phenotypic of virulence factors profile.
Collectively, our results have shown that Brazilian P. aeruginosa clinical isolates exhibit a wide variable ability to produce and express different proteases. Also, the levels of P. aeruginosa production of gelatinase and elastase were clearly isolate-specific, reaffirming that infection site and the chronicity of infection influence the virulence phenotypic of P. aeruginosa. The present study highlighted the importance of production of proteases in the physiopathology of P. aeruginosa infections, which are required for host tissue invasion and establishment of infectious process in immunosuppressed patients. In this context, the analysis of protease profile produced by P. aeruginosa clinical isolates emerges as a potential target to development of new therapeutic strategy based on virulence attenuation.
FundingThis study was supported by grants from Brazilian agencies: Fundação de Amparo à Pesquisa no Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Conflicts of interestAuthors disclose any potential financial or ethical conflicts of interest regarding the contents of this submission.
We would like to thank Denise Rocha de Souza, who is supported by a FAPERJ scholarship, for her technical assistance. The authors are in debt with Dr. Patrícia Cuervo (FIOCRUZ), who translated the abstract in order to generate its Spanish version.