The time to positivity (TTP) of blood cultures in patients with bloodstream infections (BSIs) has been considered to be a possible prognostic tool for some bacterial species. However, notable differences have been found between sampling designs and statistical methods in published studies to date, which makes it difficult to compare results or to derive reliable conclusions. Our objective was to evaluate the clinical and microbiological implications of TTP among patients with BSI caused by the most common pathogens.
MethodsA total of 361 episodes of BSI were reported for 332 patients. The survival of the entire cohort was measured from the time of blood culture sampling. In order to compare our results with those of previous studies, TTP was divided in three different groups based on log rank (short TTP <12h; medium TTP ≥12h to ≤27h, and long TTP >27h). Cox proportional hazard models were used to calculate crude and adjusted hazard ratios (HR).
ResultsThe Cox proportional hazard model revealed that TTP is an independent predictor of mortality (HR=1.00, p=0.031) in patients with BSIs. A higher mortality was found in the group of patients with the shortest TTP (<12h) (HR=2.100, p=0.047), as well as those with longest TTP (>27h) (HR=3.277, p=0.031).
ConclusionsIt seems that TTP may provide a useful prognostic tool associated with a higher risk of mortality, not only in patients with shorter TTP, but also in those with longer TTP.
El tiempo de positividad (TP) de los hemocultivos en pacientes con bacteriemia ha sido considerado como una posible herramienta pronóstica. Sin embargo, en los estudios publicados hasta la fecha, hemos observado importantes diferencias tanto en el diseño experimental como en la metodología utilizada. Esto dificulta el poder comparar los resultados obtenidos u obtener conclusiones consistentes. El objetivo de este estudio ha sido evaluar las implicaciones clínicas y microbiológicas del TP en pacientes con bacteriemia causada por los microorganismos más frecuentes, revisando la metodología utilizada en estudios anteriores.
MétodosSe estudiaron un total de 361 episodios de bacteriemia de 332 pacientes. La supervivencia de nuestra cohorte se midió desde que se tomó la muestra de hemocultivo. El TP fue dividido en tres grupos en base al log rank (TP cortos <12h; TP medios ≥12h y ≤27h; TP largos >27h), con el objetivo de comparar nuestros resultados con los obtenidos en estudios previos. Se utilizó el modelo de riesgos proporcionales (Cox) para calcular los hazard ratios (HR) tanto crudos como ajustados.
ResultadosEl modelo Cox mostró que el TP es un factor independiente relacionado con la mortalidad en pacientes con bacteriemia (HR = 1,00, p = 0,031). Concretamente, encontramos una mayor mortalidad en aquellos pacientes con TP cortos (<12 horas) (HR=2.100, p=0,047), así como en pacientes con TP largos (>27h) (HR=3.277, p=0,031).
ConclusionesEn el presente estudio demostramos que el TP puede ser utilizado como una herramienta pronóstica útil de mortalidad no solo en pacientes con TP cortos, sino también en aquellos con TP largos.
Bloodstream infections (BSIs) are a significant cause of morbidity and mortality in the general population. Over 1.2–1.4 million episodes of BSI are estimated to occur annually in Europe, leading to nearly 157,000 deaths.1 The delay on appropriate antimicrobial therapy in patients with BSI is considered a bad prognostic factor. Moreover, inappropriate use of antibiotics during BSIs is currently a global problem, increasing morbidity and mortality, provoking longer stays, higher costs and favouring development of antibiotic resistance.2 For these reasons, prompt and accurate identification of the causative pathogen is critical for guiding rapid initiation of appropriate antimicrobial therapy, diminishing mortality especially in patients with septic shock.1,2
Automated blood culture systems are the gold standard method for diagnosis of BSI in hospitals.3 These systems allow continuous monitoring of bacterial growth, which ensures faster reports to the physicians. Time to positivity (TTP) is defined as the span of time from the beginning of culture incubation to the detection of bacterial growth by an automated system. Several studies have shown that shorter TTPs are associated with a significantly higher mortality risk in patients with bacteraemia caused by several bacterial species, like Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, and Pseudomonas aeruginosa.4–7 This fact is correlated with the blood bacterial load, and therefore, shorter TTPs reflect greater disease severity.8 A majority of studies have reported significant associations between shorter TTP and risk of mortality. However, the methodology used by the different authors is dissimilar, which would decrease the reliability of the predictive models. Furthermore, the use of different statistical methods can lead to different interpretations, and could induce biased conclusions.9 The purpose of our study was to evaluate the association between TTP and mortality risk inpatients with BSIs in our hospital, examining the statistical methods carried out in previous studies in order to optimize the use of the TTP as a useful prognostic tool.
MethodsSetting and patientsA prospective study of 361 cases of BSI was carried out at the tertiary-care University Hospital Virgen del Rocío, in Seville, Spain, from January–December 2012. This study was part of an institutionally supported and educational stewardship programme for antimicrobial usage optimization, which included all clinical departments, and that has been developed in our centre in order to improve the use of antimicrobial therapy.10 This programme was launched in our hospital in the year 2011 and continues its successful development, with the first results published after one year of its implementation.11
One of the main objectives of this stewardship programme was to evaluate some indicators of its clinical impact, such as the evolution of mortality after its implementation. We performed a retrospective study of patients with microbiologically documented BSI. Since the time of diagnosis of bacteraemia due to the following organisms: E. coli, S. aureus, K. pneumoniae, Streptococcus pneumoniae, P. aeruginosa and Acinetobacter baumannii, the in-hospital mortality and the period of time until death in these patients were analyzed. In all BSI episodes that we studied, the following variables with a prognostic value were recorded: TTP (hours), sex, age, site of acquisition (nosocomial, community or health care related), source of infection, aetiology, clinical presentation (sepsis, severe sepsis, septic shock), underlying conditions (Charlson comorbidity Index) empirical antimicrobial therapy, and clinical outcome (length of stay, crude mortality and days until death). Patients under 14 years of age were excluded of this study.
Selection of articlesAll potentially relevant articles, in which TTP and mortality are related, in English and Spanish languages from January 1974 to April 2014 were identified by computerized searches of PubMed using the following Medical Subject Headings (MeSH) and keyword terms: time to positivity, bloodstream infection, sepsis, bacteraemia, blood culture and mortality. Relevant articles, judged on the basis of the title and abstract, were retrieved for more detailed evaluation.
Variable definitionsBacteremia infection was defined as ≥1 positive blood culture result with systemic manifestations of infection.12 Sepsis was defined according to the definition of the Surviving Sepsis Campaign Guidelines for Management of Severe Sepsis and Septic Shock.13 Nosocomial infection was considered when the first positive blood culture was obtained >48h after hospital admission, or when the infection occurred <48h but the patient had been hospitalized in the 3 previous weeks. Community-onset infections were considered when the first positive blood culture was obtained ≤48h after admission without previous hospitalization.14
The sources of infection were defined according to the Centres for Disease Control and Prevention (CDC) criteria.15 The diagnosis of the infection source was based on clinical, bacteriological and radiological criteria. Empirical antimicrobial therapy was defined as adequate in terms of in vitro susceptibility of an organism isolated, and if antibiotic treatment was started within 24h after drawing blood cultures. The severity of the underlying conditions was classified according to the Charlson's weighted comorbidity index.16
Microbiological methodsApproximately 20mL of blood from patients were aseptically obtained via peripheral venepuncture and distributed equally into each BACTEC™ Standard/10Aerobic/F and BACTEC™ Lytic/10 Anaerobic/F (8–10mL in each one). Samples were sent to the Microbiology laboratory and immediately incubated using a BACTEC FX system (Becton Dickinson, USA). This system monitors bacterial growth by measuring CO2 production through a fluorescent sensor every 10min along five days. Blood culture bottles were processed between 8 am and 9 pm every day, or stored during the night until 8 am.
Gram stained smears from positive bottles were visualized, and a Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Leipzig, Germany) from the sediment of the positive blood culture was directly performed as described by La Scola et al.17 The results were informed to the clinicians. The bacterial identification of all isolates were confirmed by MicroScan WalkAway®plus System (Siemens Healthcare Diagnostics, West Sacramento, CA), and MICs were determined with standard techniques according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST). TTP was defined as the span of time from the beginning of culture incubation to the detection of bacterial growth. The shortest TTP was selected for the analysis when multiple bottles were positive in the same set. Only the first positive blood culture was included in the study when the same microorganism was isolated along the hospital stay.
Statistics analysisFirst, a descriptive analysis of the documented clinical variables was carried out. Discrete variables were expressed as counts (percentages) and continuous variables as medians and interquartile range (IQR). To compare groups, the χ2 test or the Fisher's exact test were used for categorical variables, while the Mann–Whitney U or Kruskal–Wallis tests for continuous variables. We used the Wilcoxon Test with a Bonferroni-adjustment to compare TTP values among bacteria species.
The survival time of the entire cohort was measured from the time of blood culture sampling until patient's death, and was considered as the dependent variable in the multivariate Cox model. The cases that reached the endpoint of interest (death) were classified as uncensored cases, while the survivor patients or those who were discharged from the hospital before the end of the study were considered as censored cases. Median survival times were estimated by Kaplan–Meier method.18 Mantel–Cox test (Log-rank) was used to analyze univariate distributions for survival. After the cube transformation was obtained, TTP was studied as a time-dependent covariate to avoid the time-dependent bias.19 In order to compare our results with previous studies, this variable was divided in three different groups based in log rank (short TTP <12h; medium TTP ≥12h and ≤27h, and long TTP >27h). A Cox proportional hazard model20 was used to calculate crude and adjusted hazard ratios (HR) along with 95% confidence intervals (CI). Assumptions of Cox models (log-linearity, proportionality of risk in time) were met in this analysis. Because of the clinical situation variable did not meet the assumptions of proportionality of risk required for valid inference, it has been recoded as a binary variable (sepsis/severe sepsis=0, septic shock=1). The threshold for statistical significance was set at α=0.05. The free code R statistical package version 3.1.2 was used for analysis.
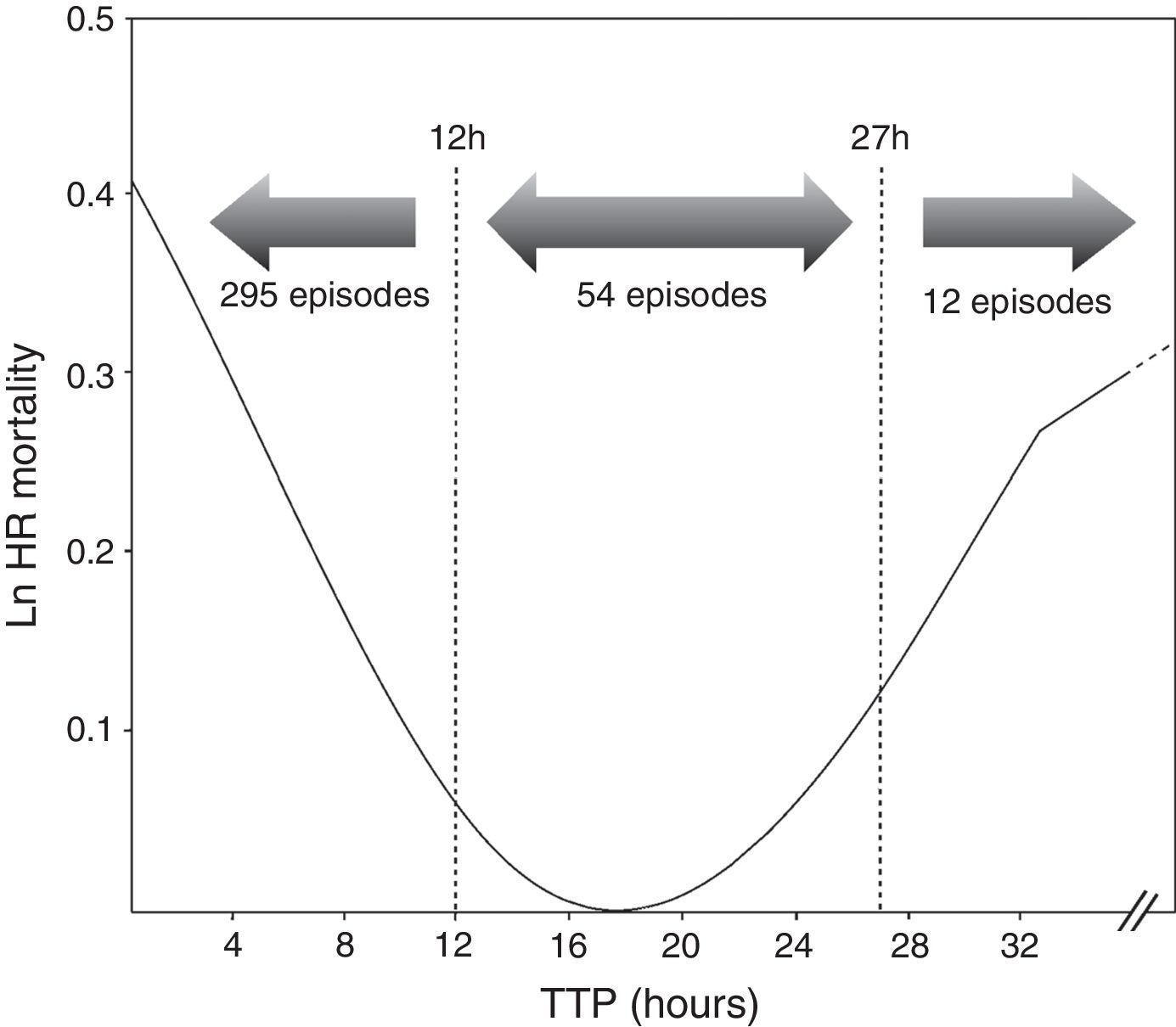
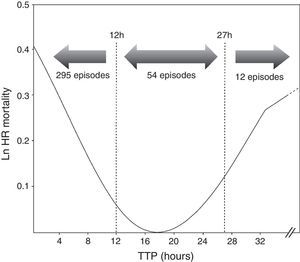
ResultsOut of 361 episodes of BSI in 332 patients aged 14–91 years, 205(56.8%) affected men and 156 (43.2%) women. The Clinical and microbiological features of the 361 episodes of BSI are shown in Table 1. Twenty-nine patients underwent a second episode of BSI. The median TTP was 7h 18min(IQR, 6h 34min). The frequency distribution of time to blood culture positivity is shown in Fig. 1. The median stay duration from blood pathogen isolation until the patient was discharged or exitus was 10 days (IQR, 16 days).
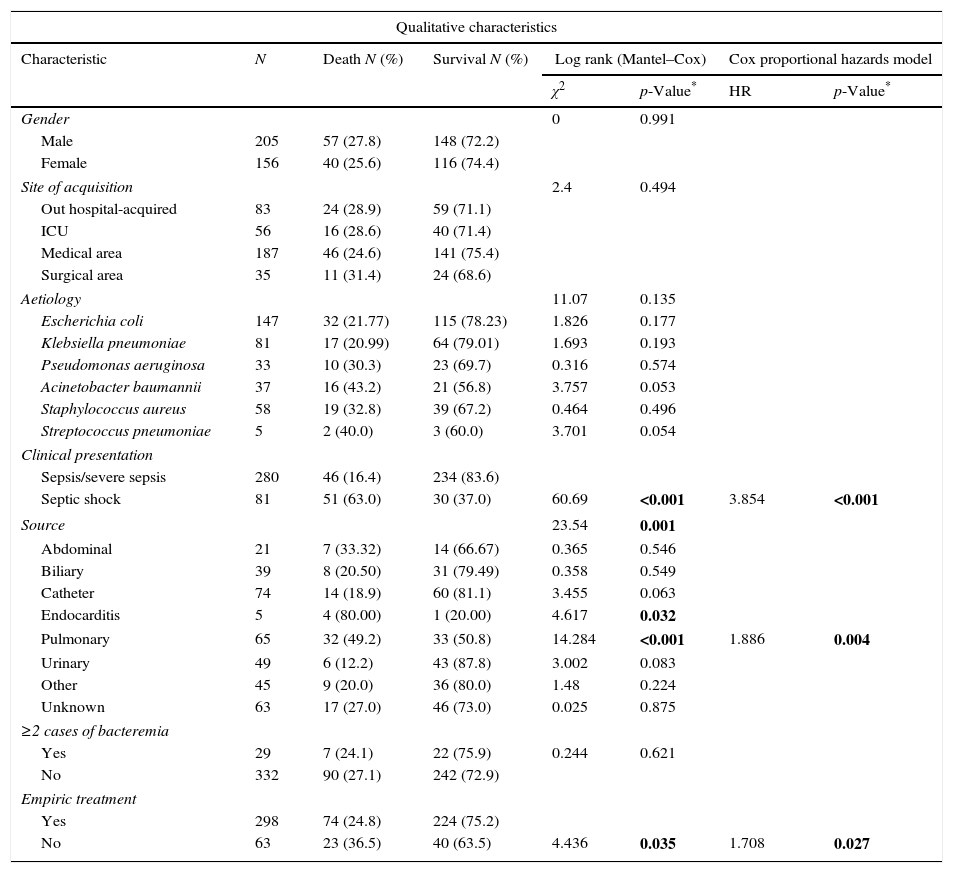
Kaplan–Meier analysis, univariate Cox and multivariate Cox proportional hazards model of clinical and microbiological characteristics of 361 BSIs episodes related with mortality included in this study.
| Qualitative characteristics | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | N | Death N (%) | Survival N (%) | Log rank (Mantel–Cox) | Cox proportional hazards model | ||
| χ2 | p-Value* | HR | p-Value* | ||||
| Gender | 0 | 0.991 | |||||
| Male | 205 | 57 (27.8) | 148 (72.2) | ||||
| Female | 156 | 40 (25.6) | 116 (74.4) | ||||
| Site of acquisition | 2.4 | 0.494 | |||||
| Out hospital-acquired | 83 | 24 (28.9) | 59 (71.1) | ||||
| ICU | 56 | 16 (28.6) | 40 (71.4) | ||||
| Medical area | 187 | 46 (24.6) | 141 (75.4) | ||||
| Surgical area | 35 | 11 (31.4) | 24 (68.6) | ||||
| Aetiology | 11.07 | 0.135 | |||||
| Escherichia coli | 147 | 32 (21.77) | 115 (78.23) | 1.826 | 0.177 | ||
| Klebsiella pneumoniae | 81 | 17 (20.99) | 64 (79.01) | 1.693 | 0.193 | ||
| Pseudomonas aeruginosa | 33 | 10 (30.3) | 23 (69.7) | 0.316 | 0.574 | ||
| Acinetobacter baumannii | 37 | 16 (43.2) | 21 (56.8) | 3.757 | 0.053 | ||
| Staphylococcus aureus | 58 | 19 (32.8) | 39 (67.2) | 0.464 | 0.496 | ||
| Streptococcus pneumoniae | 5 | 2 (40.0) | 3 (60.0) | 3.701 | 0.054 | ||
| Clinical presentation | |||||||
| Sepsis/severe sepsis | 280 | 46 (16.4) | 234 (83.6) | ||||
| Septic shock | 81 | 51 (63.0) | 30 (37.0) | 60.69 | <0.001 | 3.854 | <0.001 |
| Source | 23.54 | 0.001 | |||||
| Abdominal | 21 | 7 (33.32) | 14 (66.67) | 0.365 | 0.546 | ||
| Biliary | 39 | 8 (20.50) | 31 (79.49) | 0.358 | 0.549 | ||
| Catheter | 74 | 14 (18.9) | 60 (81.1) | 3.455 | 0.063 | ||
| Endocarditis | 5 | 4 (80.00) | 1 (20.00) | 4.617 | 0.032 | ||
| Pulmonary | 65 | 32 (49.2) | 33 (50.8) | 14.284 | <0.001 | 1.886 | 0.004 |
| Urinary | 49 | 6 (12.2) | 43 (87.8) | 3.002 | 0.083 | ||
| Other | 45 | 9 (20.0) | 36 (80.0) | 1.48 | 0.224 | ||
| Unknown | 63 | 17 (27.0) | 46 (73.0) | 0.025 | 0.875 | ||
| ≥2 cases of bacteremia | |||||||
| Yes | 29 | 7 (24.1) | 22 (75.9) | 0.244 | 0.621 | ||
| No | 332 | 90 (27.1) | 242 (72.9) | ||||
| Empiric treatment | |||||||
| Yes | 298 | 74 (24.8) | 224 (75.2) | ||||
| No | 63 | 23 (36.5) | 40 (63.5) | 4.436 | 0.035 | 1.708 | 0.027 |
| Quantitative characteristics | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | N | Death N (%) | Survival N (%) | Univariate (Cox) | Cox proportional hazards model | ||
| HR | p-Value* | HR | p-Value* | ||||
| Underlying conditions | |||||||
| Age | 1.028 | <0.001 | 1.025 | 0.001 | |||
| Charlson score index | 1.193 | <0.001 | 1.147 | <0.001 | |||
| TTPa | 1.002 | 0.002 | 1.001 | 0.031 | |||
HR, hazard ratio; ICU, intensive care unit; TTP, time to positivity.
E. coli with 147 cases (40.72%), was the most was the most frequent microorganism isolated on the BSI episodes, followed by K. pneumoniae (n=81, 22.43%), S. aureus (n=58, 16.06%), A. baumannii (n=37, 10.24%), P. aeruginosa (n=33, 9.14%), and S. pneumoniae (n=5, 1.38%). Among the 29 patients in which a second episode of BSI occurred during hospitalization, 12 were caused by E. coli (41.37%), 7 by A. baumannii (24.13%), 6 by K. pneumoniae (20.68%) and 2 by P. aeruginosa and S. aureus (6.89%). The aetiology based on the source of infection was as follows: E. coli was the most isolated microorganism for abdominal source (42.85%), biliary source (61.53%), urinary source (63.26%), unknown source (44.44%) and other sources (48.89%). S. aureus was the most isolated microorganism, when the catheter and endocarditis were the sources of infection (39.18% and 100% respectively), while A. baumannii was the most prevalent microorganism (30.76%) in the episodes with pulmonary source.
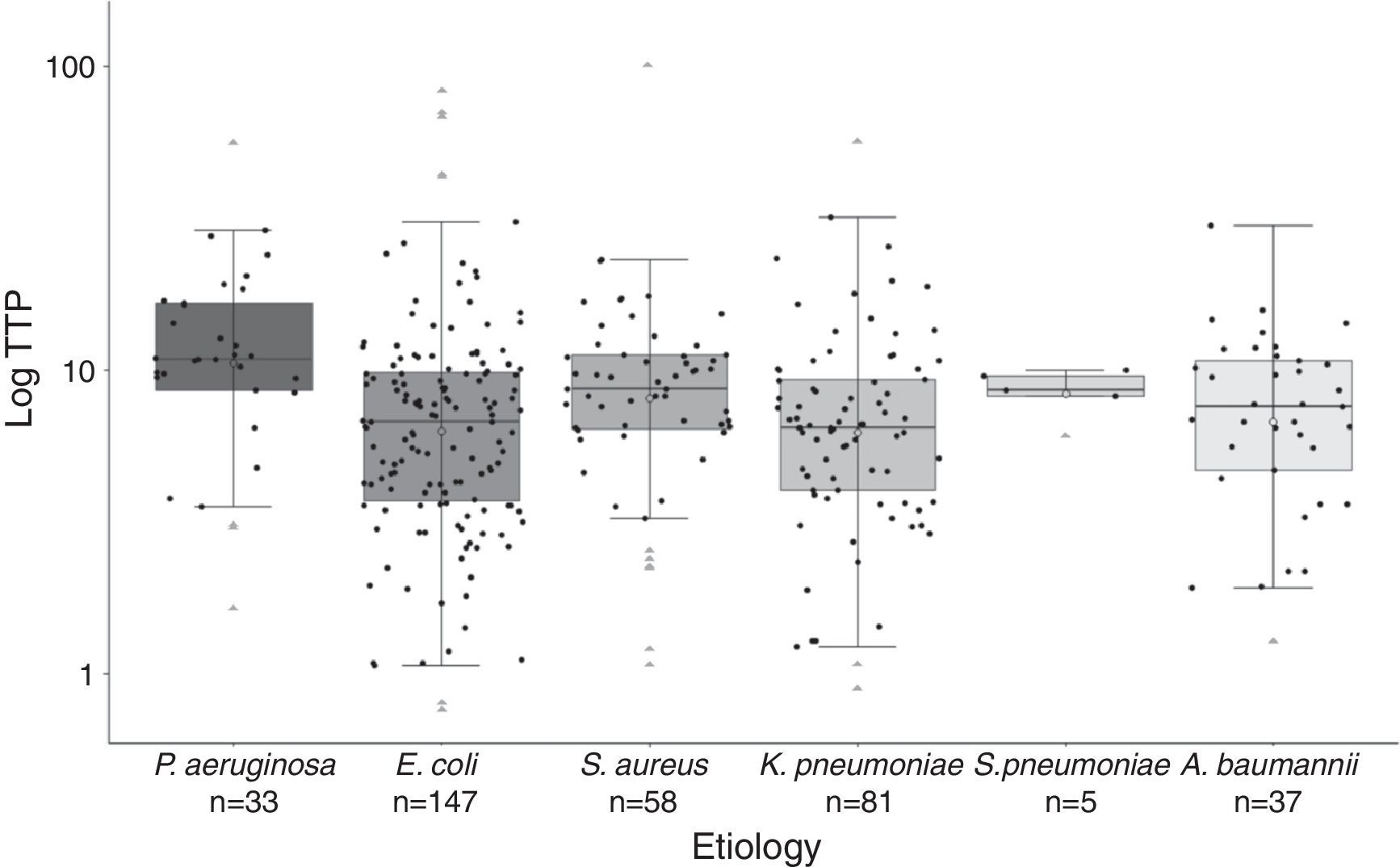
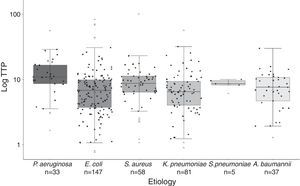
Significant differences were found in the distribution of TTP values among species (Fig. 2). Gram-negative bacilli, P. aeruginosa presented significant higher TTP values than E. coli (p<0.001) and K. penumoniae (p<0.001). No differences in TTP values between P. aeruginosa and A. baumannii were found (p=0.122). Among Gram-positive cocci, there were not significant differences between S. aureus and S. pneumoniae TTP values (p<0.99). The comparison of the TTP values between Gram-positive and Gram-negative bacteria, showed in S. aureus higher values than E. coli (p=0.043) and K. pneumoniae (p=0.026).
Boxplot graphic of the time to positivity (TTP) values for the aetiology of the 361 episodes of BSI. The distribution of each TTP value for each microorganism is recorded in this graphic (black circles). Horizontal bars inside the boxes represent medians, and empty circles represent means. The boxes represent interquartile range (IQR), and vertical bars represent values between upper and lower outlier limits. Outlier values are represented with grey triangles.
The univariate analysis (Table 1) showed a significant association among the source of infection (pulmonary source χ2=14.284, p<0.001; endocarditis χ2=4.617, p=0.032), clinical presentation (Septic shock, χ2=60.69, p<0.001) and empirical antimicrobial treatment (yes vs. no χ2=4.436, p=0.035) with mortality risk. The univariate Cox model for continuous variables showed significant associations among age (HR=1.004, p=0.006) and Charlson comorbidity index (HR=1.205, p<0.001) with mortality risk.
Cox proportional hazard model assessed TTP as an independent predictor of mortality (HR=1.001, p=0.031). All variables included in the final Cox model are shown in Table 1. Additionally, two cut-off values were evaluated to compare the results obtained with previous studies. A higher mortality was found in the group of patients with the shortest TTP (<12h) (HR=2.100, p=0.047) as well as those with longest TTP (>27h) (HR=3.277, p=0.031) (Table 2). Comparing the clinical characteristics of patients with short, medium and long TTP values, we found significant differences in the source of infection, being more common the abdominal source in patients with long TTP (23.1% of total) compared with those with medium TTP values (3.8%), and short TTP values (5.4%) (χ2=7.56, p=0.023). Moreover, a higher rate of BSI caused by P. aeruginosa were found in patients with longer TTP (23.1%) compared with medium (18.9%) and short (6.8%) TTP values (χ2=11.06, p=0.004).
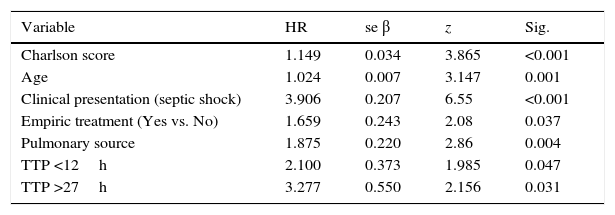
Summarize of Cox regression model with all variables identified in the bivariate analysis, studying the TTP as a categorized variable in order to compare with previous studies.
| Variable | HR | se β | z | Sig. |
|---|---|---|---|---|
| Charlson score | 1.149 | 0.034 | 3.865 | <0.001 |
| Age | 1.024 | 0.007 | 3.147 | 0.001 |
| Clinical presentation (septic shock) | 3.906 | 0.207 | 6.55 | <0.001 |
| Empiric treatment (Yes vs. No) | 1.659 | 0.243 | 2.08 | 0.037 |
| Pulmonary source | 1.875 | 0.220 | 2.86 | 0.004 |
| TTP <12h | 2.100 | 0.373 | 1.985 | 0.047 |
| TTP >27h | 3.277 | 0.550 | 2.156 | 0.031 |
HR, hazard ratio; se β, regression coefficient; z, dichotomous indicator; TTP, time to positivity.
Additional independent risk factors correlated with mortality in the final adjusted model were: Charlson comorbidity index (HR=1.149, p<0.001); clinical presentation (septic shock, HR=3.91, p<0.001); age (HR=1.025, p=0.001); empirical therapy (not treatment, HR=1.66, p=0.037); and pulmonary source of infection (HR=1.87, p=0.004) (Table 2).
DiscussionIn the present study we demonstrate that, in patients with BSI caused by the most common pathogens, the TTP may be used as a useful prognostic tool for clinical management. To our knowledge, only one previous study has associated longer TTP values with increased mortality risk in patients with BSI, and only when S. aureus was involved. Thus, our study is the first describing this association with both Gram-negative and Gram-positive bacteria, also analyzing the associated clinical and microbiological factors. We also present an appropriate methodology to avoid biased estimates and subsequent flawed interpretations of the data.
Previous studies have suggested that blood bacterial concentration could be related to clinical outcome in patients with BSI.21 In Clinical Microbiology laboratories, the TTP of blood cultures are automatically and continuously monitored, giving to microbiologists a potential marker of bacterial load in blood. Several authors have related the higher bacterial load in blood with shorter TTPs and mortality. Thus, shorter TTP values could reflect a more severe bacteremia, and could be associated with a higher probability of complications and death.4,22 Therefore, TTP appears to provide useful diagnostic and prognostic information, and should be considered when risk of death in patients with BSI is evaluated.
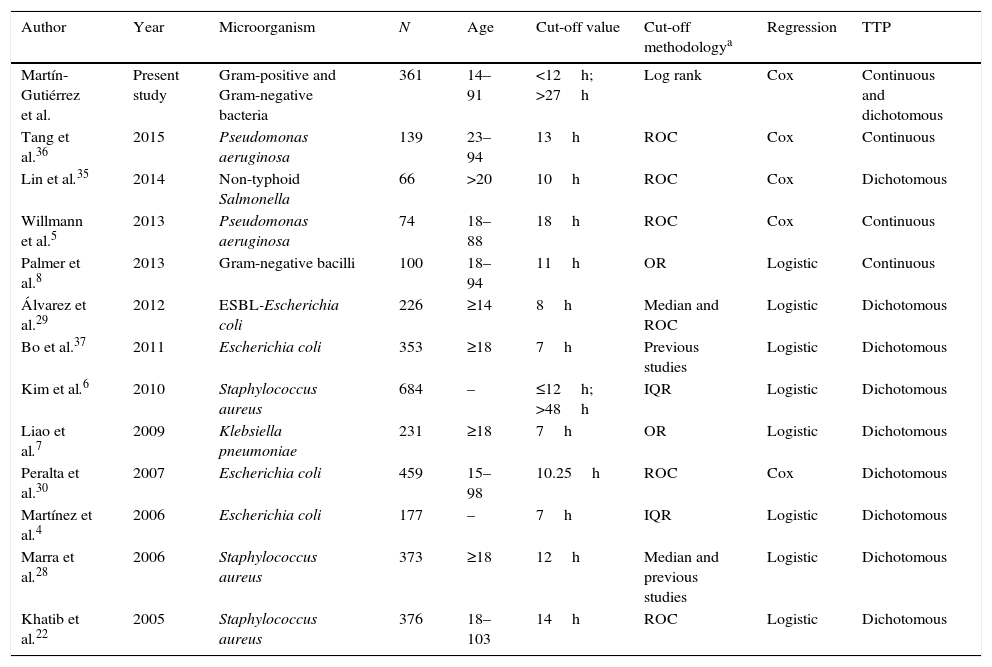
The results of our study agree with previous studies where the TTP of blood cultures is correlated with the clinical outcome of patients. Twelve studies, using different cut-off values, and which have analyzed this correlation, have been reviewed. In all of them a multivariate analysis was performed, but only in four a Cox regression model was carried out (Table 3). When evaluation of the relationship between survival time until the occurrence of an event (in our case death) with one or more predictors is required in the epidemiological studies, the most recommendable and widely used method is the Cox proportional-hazards regression model. Moreover, the practice of dichotomizing continuous covariates is common in medical and epidemiological researches.23 The categorization of the prognostic variables is performed in order to set up practical eligibility criteria, with a simple risk classification of higher risk vs. lower risk, allowing to select the so called best threshold value for a covariate.24,25 The methodology used in previous reports to select the TTP optimum cut-point is widely diverse, such as the median, IQR, ROC, or OR values. The dichotomization of a continuous covariate cannot be arbitrary, because this would affect its usefulness when assessing the true prognostic value of a variable.26,27 In survival analyses, the methods which are expected to have better statistical power are those based on log rank score and likelihood ratio.25 Furthermore, when a variable is categorized from a continuous one, there is always the possibility of losing information as well as statistical power to reach significance.24
Summary of previous studies assessing the correlation between TTP and risk of mortality.
| Author | Year | Microorganism | N | Age | Cut-off value | Cut-off methodologya | Regression | TTP |
|---|---|---|---|---|---|---|---|---|
| Martín-Gutiérrez et al. | Present study | Gram-positive and Gram-negative bacteria | 361 | 14–91 | <12h; >27h | Log rank | Cox | Continuous and dichotomous |
| Tang et al.36 | 2015 | Pseudomonas aeruginosa | 139 | 23–94 | 13h | ROC | Cox | Continuous |
| Lin et al.35 | 2014 | Non-typhoid Salmonella | 66 | >20 | 10h | ROC | Cox | Dichotomous |
| Willmann et al.5 | 2013 | Pseudomonas aeruginosa | 74 | 18–88 | 18h | ROC | Cox | Continuous |
| Palmer et al.8 | 2013 | Gram-negative bacilli | 100 | 18–94 | 11h | OR | Logistic | Continuous |
| Álvarez et al.29 | 2012 | ESBL-Escherichia coli | 226 | ≥14 | 8h | Median and ROC | Logistic | Dichotomous |
| Bo et al.37 | 2011 | Escherichia coli | 353 | ≥18 | 7h | Previous studies | Logistic | Dichotomous |
| Kim et al.6 | 2010 | Staphylococcus aureus | 684 | – | ≤12h; >48h | IQR | Logistic | Dichotomous |
| Liao et al.7 | 2009 | Klebsiella pneumoniae | 231 | ≥18 | 7h | OR | Logistic | Dichotomous |
| Peralta et al.30 | 2007 | Escherichia coli | 459 | 15–98 | 10.25h | ROC | Cox | Dichotomous |
| Martínez et al.4 | 2006 | Escherichia coli | 177 | – | 7h | IQR | Logistic | Dichotomous |
| Marra et al.28 | 2006 | Staphylococcus aureus | 373 | ≥18 | 12h | Median and previous studies | Logistic | Dichotomous |
| Khatib et al.22 | 2005 | Staphylococcus aureus | 376 | 18–103 | 14h | ROC | Logistic | Dichotomous |
TTP, time to positivity; ROC, receiver operating characteristic; OR, odds ratio; IQR, interquartilic range.
In our study, TTP values of blood cultures were considered to be a continuous covariate, as in Willmann et al.5 and Palmer et al.8 Additionally, we decided to analyze the TTP in three separate groups based on the log rank to compare our results with previous studies. We found that TTP was not a linear-related risk of mortality, showing a higher risk the patients with the shortest TTP values (<12h), as well as the patients with the longest TTP values (>27h) (Fig. 1). The association between shorter TTP values and increased mortality has been reported in previous studies.22,28–30 However, the association between the longer TTP values and the mortality increase was only found in patients with S. aureus BSI by Kim et al.6 Non-significant differences were found on clinical characteristics of patients with short and long TTP values, suggesting the authors that correlation of the delayed TTP and mortality could be blamed on the low-grade bacteremia, which is not accompanied with severe clinical manifestations, this often leading to delayed or inappropriate antimicrobial treatment and consequently a worst clinical outcome.
Here, we have found significant differences in anatomical source of infection between patients with long TTP and those with intermediate or short TTP. In particular, the abdominal source of infection was more common in patients with long TTP (23.1% of total) than in those with intermediate (3.8%) or low TTP values (5.4%). Additionally, the number of patients with BSI caused by P. aeruginosa was significantly higher in longer TTP group (23.1%) compared with medium (18.9%) and short (6.8%) TTP values. Previous studies have shown that P. aeruginosa BSI is a bad prognostic factor, remaining fatal in more than 20% of the cases.31,32 Moreover, P. aeruginosa is intrinsically resistant to many antibiotics, exhibiting an inherently reduced susceptibility to antibiotics compared with other Gram-negative bacteria, and it has been related with antimicrobial treatment failures.33,34
All age groups and the six most frequent microorganisms of BSI are included in our cohort, reflecting both circumstances the daily clinical practice in our Hospital. Thus, as previously published by Martínez et al.4 and Palmer et al.,8 lactose-fermenting Gram-negative bacilli presented higher TTP values compared with non-lactose fermenters. P. aeruginosa presented significant higher TTP values compared with K. penumoniae and E. coli. Gram-positive cocci also presented higher TTP values compared with lactose-fermenting Gram-negative bacilli (Fig. 2). Although we found significant differences in the TTP values by aetiology, this covariate was not actually related to deleterious outcome.
The remaining independent variables, significantly related to mortality and included in the final Cox regression model, support previous findings. Thus, the association of higher mortality in patients with septic shock was described.7,30,35 The presence of underlying condition assessed by the Charlson Index was also related with higher mortality.6,28,29 The initiation of empiric treatment was an independent protective factor for mortality in bacteremic infections, as previously shown by Willmann et al.5 The respiratory source of infection was significantly associated with mortality compared with others sources of infection, as described by Kim et al.4 and Martinez et al.6 Although endocarditis as a source of bacteraemia has been related with mortality,22 it has been dropped out from our final model, probably due to the low number of patients with endocarditis recorded in our study: only five patients (four of them died).
The present study has several limitations such as the timing of blood sample collection was not measured, nor the exact quantity of blood inoculated into the BACTEC™, and both facts may affect the final TTP values. Moreover, some clinical characteristics are not considered in this study, such as immune status of patients or previous surgery processes, and may act as confounding variables in survival studies. We neither included the appropriate antibiotic treatment in the final statistical analysis because there were several cases with incomplete information.
The aim of this paper was to study the association between TTP and mortality in patients with BSI using the most appropriate statistical methods, in order to produce unbiased estimates and fitting p values. In this way, we found that TTP may provide a useful prognostic tool associated with higher risk of mortality not only in patients with shorter TTP, but also in those with longer TTP. Further prospective studies are needed in order to contrast these data in larger populations, mainly in patients with longer TTP values.
Funding informationThis research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interestsThe authors declare no conflict of interest.