Asthma is a complex disease determined by the interaction of different genes and environmental factors. The first genetic investigations in asthma were candidate gene association studies and linkage studies. In recent years research has focused on association studies that scan the entire genome without any prior conditioning hypothesis: the so-called genome-wide association studies (GWAS). The first GWAS was published in 2007, and described a new locus associated to asthma in chromosome 17q12-q21, involving the ORMDL3, GSDMB and ZPBP2 genes (a description of the genes named in the manuscript are listed in Table 1). None of these genes would have been selected in a classical genetic association study since it was not known they could be implicated in asthma. To date, a number of GWAS studies in asthma have been made, with the identification of about 1000 candidate genes. Coordination of the different research groups in international consortiums and the application of new technologies such as new generation sequencing will help discover new implicated genes and improve our understanding of the molecular mechanisms underlying the disease.
Asthma is clinically characterised by bronchial hyperresponsiveness associated to airway obstruction episodes. Bronchial obstruction is the result of an inflammatory process triggered by allergens, infections or other still little known factors. The prevalence of the disease is high, particularly in developed countries. In the last few decades there has been a notorious increase in the number of cases of asthma. Although it has been known for a long time that asthma has an important hereditary component, the rapidness with which the prevalence of the disease has increased suggests that not only genetic factors are involved. In this respect it has been suggested that certain environmental factors, particularly related to the industrialisation process, and their influence on the genetic component of the disease, could contribute to increasing the frequency of allergic diseases in general, and of asthma in particular. However, in recent decades the identification of specific genes associated to the risk of suffering the disease, and their validation, has been one of the challenges facing research in asthma. The Human Genome Project is an international initiative designed to identify the sequence of the human genome. Following the publication in 2001 of a draft of the sequence, investigators have focused on genomic variations among individuals. In this respect, polymorphisms are variations in the genetic sequence that are found in over 1% of the population. Single nucleotide polymorphisms (SNPs) are polymorphisms that affect a single base. It has been estimated that as of 23 July 2013, the single nucleotide polymorphism database (dbSNP) listed 62,676,337 SNPs in humans (http://www.ncbi.nlm.nih.gov/mailman/pipermail/dbsnp-announce/2013q3/000133.html). Some of these variations could affect the quantity, timing and function of a protein encoded by a gene, and contribute to the risk of suffering a disease. Most diseases are caused by an interaction between genetic and environmental factors.1 Many case–control studies have been carried out to clarify the relationship between genetic variations and specific diseases.2
The present review analyses how GWAS can contribute to improving our understanding of asthma through the identification of markers specifically related to the disease (Table 1).
Description of genes by alphabetical order.
| Name/gene ID | Description |
|---|---|
| AGER | Advanced glycosylation end product-specific receptor |
| C11orf30 | Chromosome 11 open reading frame 30 |
| CD14 | CD14 molecule |
| CHRNA3/5 | Cholinergic receptor, nicotinic, alpha 3/5 |
| CRB1 | Crumbs family member 1, photoreceptor morphogenesis associated |
| CRIM1 | Cysteine rich transmembrane BMP regulator 1 |
| DENND1B | DENN/MADD domain containing 1B |
| FAM13A | Family with sequence similarity 13, member A |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| FOXP3 | Forkhead box P3 |
| GAB1 | GRB2-associated binding protein 1 |
| GATA3 | GATA binding protein 3 |
| GLCCI1 | Glucocorticoid induced transcript 1 |
| GSDMB | Gasdermin B |
| GSTCD | Glutathione S-transferase, C-terminal domain containing |
| HHIP | Hedgehog interacting protein |
| HLA-DQ | Major histocompatibility complex, class II, DQ |
| IKZF4 | IKAROS family zinc finger 4 |
| IL1RL1 | Interleukin 1 receptor-like 1/ |
| IL2RB | Interleukin 2 receptor beta |
| IL-3 | Interleukin 3 |
| IL4R | Interleukin 4 receptor |
| IL-6 | Interleukin 6 |
| IL6R | Interleukin 6 receptor |
| IL12A, | Interleukin 12A |
| IL12RB1 | Interleukin 12 receptor, beta 1 |
| IL13 | Interleukin 13 |
| IL18R1 | Interleukin 18 receptor 1 |
| IL33 | Interleukin 33 |
| IRF2 | Interferon regulatory factor 2 |
| LOC730217 | Hypothetical LOC730217 |
| LRRC32 | Leucine rich repeat containing 32 |
| MYB | V-myb avian myeloblastosis viral oncogene homolog |
| NOTCH4 | Notch 4 |
| OPRK1 | Opioid receptor, kappa 1 |
| OR7E156P | Olfactory receptor, family 7, subfamily E, member 156 pseudogene |
| ORMDL3 | ORM1-like 3 |
| PDCD4 | Programmed Death Cell Domain 4 |
| PDE4D | Phosphodiesterase 4D, cAMP-specific |
| PDE11A | Phosphodiesterase 11A |
| PRKG1 | Protein kinase, cGMP-dependent, type I |
| PTGDR | Prostaglandin D2 Receptor Gene |
| PYHIN1 | Pyrin and HIN domain family, member 1 |
| RAD50 | RAD50 homolog |
| RARB | Retinoic acid receptor, beta |
| RPL32P28 | Ribosomal protein L32 pseudogene 28p |
| SMAD3 | SMAD family member 3 |
| STAT4 | Signal transducer and activator of transcription 4 |
| STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced |
| SYNPO2 | Synaptopodin 2 |
| THSD4 | Thrombospondin, type I, domain containing 4 |
| TLE4 | Transducin-like enhancer of split 4 |
| TLN1 | Talin 1 |
| TNFR1 | Tumour necrosis factor receptor 1 |
| TSLP | Thymic stromal lymphopoietin |
| USP38 | Ubiquitin-specific peptidase 38 |
| ZNF323 | Zinc finger protein 323 |
| ZPBP2 | Zona pellucida binding protein 2 |
Genome-wide association studies (GWAS) allow the analysis of hundreds of thousands of polymorphisms with the purpose of relating them to a specific phenotype, generally a disease such as asthma, or a characteristic of the disease such as the levels of IgE. The main advantage of GWAS has been their excellent resolution, their statistical power in detecting risk variants with moderate effects, and their capacity to analyse the entire genome. GWAS has offered the opportunity to identify new genes or regulatory regions not previously related to the disease, but without the need to study families. They are able to detect loci with modest effects upon the disease, although GWAS are not useful for detecting rare disease variants.
The inconveniences of GWAS are that they need large sample sizes and tools for processing a very large body of data. Because of the great number of statistical comparisons in GWAS (hundreds of thousands), the threshold defining significant associations is different from that of conventional case-control studies. The most restrictive threshold is p<7.2×10−8, and the least restrictive p<5×10−7.2 Many of the GWAS published to date have not yielded loci with the level of statistical significance required to indicate a genuine association. As a result, these studies have not fully met the initial expectations, although the information produced may be very useful.
Large international research consortiums have been founded with the aim of conducting rigorous statistical GWAS meta-analyses for common diseases. In Europe, the GABRIEL consortium was created for the study of asthma, analysing a population of 10,365 cases and 16,110 controls.4 The European initiative was followed by the EVE consortium comprising North American investigators. The EVE consortium fundamentally reflects the ethnic diversity of North America, comprising a population of European, African/Caribbean and Hispanic origin, with the inclusion of over 5000 cases of asthma.5 At present, the Transnational Asthma Genetics Consortium (TAGC) is conducting an international GWAS that will allow the completion of a meta-analysis of asthma with the examination of about 30,000 cases and 50,000 controls.6
GWAS in asthmaThe first GWAS in asthma was carried out in 2007 with a population of approximately 1000 cases and 1000 controls.7 It identified the ORMDL3 gene in chromosome 17q21 as a contributor to the risk of developing childhood asthma. This finding has been recognised as the most consistent genetic association obtained by a GWAS in asthma to date. Genetic variants in this particular locus regulate expression of the OMRDL3 gene and the neighbouring GSDML gene in asthmatic children. The variant rs7216389-T, which shows an increased association to asthma and increased expression of the OMRDL3 gene, is located in a region with high linkage disequilibrium in chromosome 17q21, very close to the ORMDL3 gene and within the first intron of the GSDML gene. This non-coding region shows notorious homology among species and contains a possible binding element for the transcription factor C/EBPβ, which is very important in regulating the genes implicated in immune and inflammatory responses.7 Due to the important degree of linkage among the SNPs and the co-regulation of genic expression in this region, it has not been possible to distinguish between the effects of these two genes; we therefore generally speak of an association to asthma on the part of locus 17q21.2 This association has been reproduced in several studies in different ethnic populations, though it seems that the mentioned genomic region is associated to childhood asthma, but not to adult asthma.2,3 Sleiman et al.8 carried out a GWAS in children with persistent asthma requiring daily inhaled glucocorticoid therapy, and a control group. In addition to confirming the association to locus 17q21, this study identified a new locus related to asthma susceptibility in chromosome 1q31, which contains the CRB1 and DENND1B genes. DENND1B encodes for a protein with a TNFR1 binding domain. It constitutes a negative TNFR1 regulator in response to cytokine-induced stress. DENND1B is expressed both by dendritic cells and by activated T cells, and modulates the Th1-Th2 cytokine cascade and other inflammatory signals through the repression of TNFR1.8
A GWAS carried out in a paediatric population in North America identified a strong association to the PDE4D gene (5q12), which encodes for a bronchial phosphodiesterase. The association was not observed in children of African origin.3 Belonging to the same family as PDE4D, the PDE11A gene was associated to asthma in a GWAS involving six-year-old asthmatic children. Although statistical significance was not reached, it seems that this phosphodiesterase family may play a role in the pathogenesis of asthma.3 Another GWAS found an association to the TLE4 gene (9q21) in a population of 492 Mexican children – although statistical significance was likewise not reached in this case. Nevertheless, this gene was replicated in a Mexican cohort. The mentioned gene, which participates in the development of B lymphocytes, had not been previously associated to asthma.3
The meta-analysis carried out by the GABRIEL consortium identified six susceptible loci, containing the IL1RL1/IL18R1, HLA-DQ, IL33, SMAD3, ORMDL3/GSDMB and IL2RB genes, which participate in Th1 or Th2 cell responses.3,4
In 2011, the EVE consortium conducted a meta-analysis of GWAS in asthma among North American populations.5 A total of five susceptibility loci were identified: locus 17q21 (GSDMB/ORMDL3) and the previously described IL1RL1, TSLP and IL33 genes.4 In addition, a new asthma-related locus specific of individuals of African origin was identified in the PYHIN1 gene.
A GWAS in asthma was carried out in 2011 in a Japanese population of adults, with the description of five candidate loci.6 Association of the HLA locus (6p21) was confirmed, with identification of the TSLP gene (5q22), which participates in the maturation of Th2 lymphocytes through the activation of antigen presenting cells (APCs). This locus had already been associated by Torgerson in a North American population.5 The locus containing the USP38-GAB1 genes (4q31) was also described. USP38 encodes for ubiquitin-specific peptidase 38 (USP38), the function of which remains uncertain. GAB1 in turn is a protein that participates in signalling pathways activated by the receptors of cytokines IL-3, IL-6, Interferon α and γ, and the B and T lymphocyte receptors. A region was identified in chromosome 10 that does not appear to contain a coding gene, but which is located at 1Mb in 3′ sense from the GATA3 gene, the main regulator of Th2 lymphocyte differentiation. In chromosome 12q13 is located IKZF4 gene (also known as EOS) that is involved in the differentiation of regulatory T lymphocytes as a FOXP3 silencing co-regulator.6
A GWAS was carried out in an Australian population in combination with a large number of samples from the GABRIEL consortium. Significant associations were observed in the IL6R (1q21), C11orf30/LRRC32 (11q13), PRKG1 (10q11) and RPL32P28/OR7E156P (13q21) genes.3
Approximately 5% of all asthma patients suffer a serious form of the disease that is difficult to control with the existing treatments. Li et al.6 focused on this kind of patients in their GWAS. A total of 292,443 SNPs were studied in a population of cases supplied by the Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR), versus a control population. The study confirmed the relevance of the regions corresponding to the RAD50-IL13 (5q31 and HLA-DR/DQ genes (6p21), implicated in the regulation of Th2 cytokines and in antigen presentation in difficult to control severe asthma.
GWAS have also been used in pharmacogenetic studies. In this respect, a recent publication has identified genic variants associated to changes in lung function in response to inhaled corticosteroids. A functional variant of GLCCI1 was associated to an important decrease in patient response to corticosteroid treatment.6
Immunoglobulin E levels are closely correlated to the clinical expression of allergic asthma. Several GWAS have found variants that may modulate IgE levels in asthmatic individuals, including FCER1A, STAT6, IL4R, IL13 and HLA.3 A recent GWAS in a Korean population of 877 asthmatic subjects has identified new variants in the CRIM1, TLN1 and SYNPO2 genes, associated to total IgE levels. In addition, in this group of asthmatics, an association was found in the intergenic region close to the OPRK1 and LOC730217 genes correlated to the levels of specific IgE against the dust mites Dermatophagoides pteronyssinus and D. farinae, respectively.9
A meta-analysis was carried out in 2012 using data from 19 GWAS, with a total of about 23,000 individuals mostly of European origin, of whom 8165 were asthmatics. The aim of the study was to identify genic variants associated to body mass index (BMI) in asthmatic children and adults.10 Associations were found between variants of the DENND1B gene and BMI in the asthmatic children. However, this association could not be replicated in independent groups, and the effect was moreover heterogeneous depending on the geographical setting considered. The mentioned gene had already been related to the risk of suffering asthma in children,8 and the detection of an association to another phenotype is interesting – although replication of the association is needed in order to validate the results.
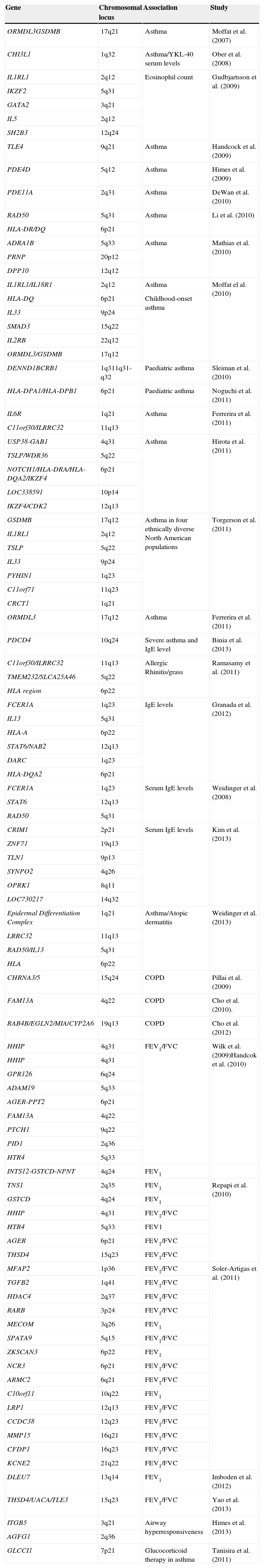
Altered lung function is a phenotype associated to asthma and to chronic obstructive pulmonary disease. Variants of the HHIP and CHRNA3/5 genes have been associated to this phenotype and have been replicated in several populations.3Table 2 enumerates other loci associated to lung function.
Published genome-wide association studies of asthma and related phenotypes.
| Gene | Chromosomal locus | Association | Study |
|---|---|---|---|
| ORMDL3GSDMB | 17q21 | Asthma | Moffat et al. (2007) |
| CHI3L1 | 1q32 | Asthma/YKL-40 serum levels | Ober et al. (2008) |
| IL1RL1 | 2q12 | Eosinophil count | Gudbjartsson et al. (2009) |
| IKZF2 | 5q31 | ||
| GATA2 | 3q21 | ||
| IL5 | 2q12 | ||
| SH2B3 | 12q24 | ||
| TLE4 | 9q21 | Asthma | Handcock et al. (2009) |
| PDE4D | 5q12 | Asthma | Himes et al. (2009) |
| PDE11A | 2q31 | Asthma | DeWan et al. (2010) |
| RAD50 | 5q31 | Asthma | Li et al. (2010) |
| HLA-DR/DQ | 6p21 | ||
| ADRA1B | 5q33 | Asthma | Mathias et al. (2010) |
| PRNP | 20p12 | ||
| DPP10 | 12q12 | ||
| IL1RL1/IL18R1 | 2q12 | Asthma | Moffat el al. (2010) |
| HLA-DQ | 6p21 | Childhood-onset asthma | |
| IL33 | 9p24 | ||
| SMAD3 | 15q22 | ||
| IL2RB | 22q12 | ||
| ORMDL3/GSDMB | 17q12 | ||
| DENND1BCRB1 | 1q311q31-q32 | Paediatric asthma | Sleiman et al. (2010) |
| HLA-DPA1/HLA-DPB1 | 6p21 | Paediatric asthma | Noguchi et al. (2011) |
| IL6R | 1q21 | Asthma | Ferrerira et al. (2011) |
| C11orf30/ILRRC32 | 11q13 | ||
| USP38-GAB1 | 4q31 | Asthma | Hirota et al. (2011) |
| TSLP/WDR36 | 5q22 | ||
| NOTCH1/HLA-DRA/HLA-DQA2/IKZF4 | 6p21 | ||
| LOC338591 | 10p14 | ||
| IKZF4/CDK2 | 12q13 | ||
| GSDMB | 17q12 | Asthma in four ethnically diverse North American populations | Torgerson et al. (2011) |
| IL1RL1 | 2q12 | ||
| TSLP | 5q22 | ||
| IL33 | 9p24 | ||
| PYHIN1 | 1q23 | ||
| C11orf71 | 11q23 | ||
| CRCT1 | 1q21 | ||
| ORMDL3 | 17q12 | Asthma | Ferrerira et al. (2011) |
| PDCD4 | 10q24 | Severe asthma and IgE level | Binia et al. (2013) |
| C11orf30/ILRRC32 | 11q13 | Allergic Rhinitis/grass | Ramasamy et al. (2011) |
| TMEM232/SLCA25A46 | 5q22 | ||
| HLA region | 6p22 | ||
| FCER1A | 1q23 | IgE levels | Granada et al. (2012) |
| IL13 | 5q31 | ||
| HLA-A | 6p22 | ||
| STAT6/NAB2 | 12q13 | ||
| DARC | 1q23 | ||
| HLA-DQA2 | 6p21 | ||
| FCER1A | 1q23 | Serum IgE levels | Weidinger et al. (2008) |
| STAT6 | 12q13 | ||
| RAD50 | 5q31 | ||
| CRIM1 | 2p21 | Serum IgE levels | Kim et al. (2013) |
| ZNF71 | 19q13 | ||
| TLN1 | 9p13 | ||
| SYNPO2 | 4q26 | ||
| OPRK1 | 8q11 | ||
| LOC730217 | 14q32 | ||
| Epidermal Differentiation Complex | 1q21 | Asthma/Atopic dermatitis | Weidinger et al. (2013) |
| LRRC32 | 11q13 | ||
| RAD50/IL13 | 5q31 | ||
| HLA | 6p22 | ||
| CHRNA3/5 | 15q24 | COPD | Pillai et al. (2009) |
| FAM13A | 4q22 | COPD | Cho et al. (2010). |
| RAB4B/EGLN2/MIA/CYP2A6 | 19q13 | COPD | Cho et al. (2012) |
| HHIP | 4q31 | FEV1/FVC | Wilk et al. (2009)Handcok et al. (2010) |
| HHIP | 4q31 | ||
| GPR126 | 6q24 | ||
| ADAM19 | 5q33 | ||
| AGER-PPT2 | 6p21 | ||
| FAM13A | 4q22 | ||
| PTCH1 | 9q22 | ||
| PID1 | 2q36 | ||
| HTR4 | 5q33 | ||
| INTS12-GSTCD-NPNT | 4q24 | FEV1 | |
| TNS1 | 2q35 | FEV1 | Repapi et al. (2010) |
| GSTCD | 4q24 | FEV1 | |
| HHIP | 4q31 | FEV1/FVC | |
| HTR4 | 5q33 | FEV1 | |
| AGER | 6p21 | FEV1/FVC | |
| THSD4 | 15q23 | FEV1/FVC | |
| MFAP2 | 1p36 | FEV1/FVC | Soler-Artigas et al. (2011) |
| TGFB2 | 1q41 | FEV1/FVC | |
| HDAC4 | 2q37 | FEV1/FVC | |
| RARB | 3p24 | FEV1/FVC | |
| MECOM | 3q26 | FEV1 | |
| SPATA9 | 5q15 | FEV1/FVC | |
| ZKSCAN3 | 6p22 | FEV1 | |
| NCR3 | 6p21 | FEV1/FVC | |
| ARMC2 | 6q21 | FEV1/FVC | |
| C10orf11 | 10q22 | FEV1 | |
| LRP1 | 12q13 | FEV1/FVC | |
| CCDC38 | 12q23 | FEV1/FVC | |
| MMP15 | 16q21 | FEV1/FVC | |
| CFDP1 | 16q23 | FEV1/FVC | |
| KCNE2 | 21q22 | FEV1/FVC | |
| DLEU7 | 13q14 | FEV1 | Imboden et al. (2012) |
| THSD4/UACA/TLE3 | 15q23 | FEV1/FVC | Yao et al. (2013) |
| ITGB5 | 3q21 | Airway hyperresponsiveness | Himes et al. (2013) |
| AGFG1 | 2q36 | ||
| GLCCI1 | 7p21 | Glucocorticoid therapy in asthma | Tanisira et al. (2011) |
Abbreviations: COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1s; FVC, forced vital capacity; IgE, immunoglobulin E.
In recent months a number of studies have conducted in depth evaluations of previously published GWAS. Li et al. investigated the association of genic variants related to lung function in four North American populations of asthmatic Caucasians based on GWAS and meta-analyses. Four of the 32 loci associated to impaired lung function corresponded to type Th1 genes (IL12A, IL12RB1, STAT4 and IRF2).11
Two previous meta-analyses in the general population found 28 loci associated to the regulation of lung function. The meta-analysis conducted in asthmatics identified seven of these 28 loci (HHIP, FAM13A, THSD4, GSTCD, NOTCH4-AGER, RARB and ZNF323), fundamentally implicated in airway structure and remodelling.11
On the other hand, no association of type Th2 genes (which confer susceptibility to asthma) (IL13, TSLP, IL33 and IL1RL1) to lung function was found – thus suggesting that the genes associated to lung function which might influence the severity of asthma are different from the genes associated to asthma susceptibility. The authors proposed a genetic model of asthma progression in which genic variants in type Th2 genes induce a Th2 dominant atopic and asthma susceptibility response. Genetic variants in type Th1 genes or IL-12, and in airway structure and remodelling genes, together with other mechanisms, affect lung function and increase the severity of asthma.11
Binia et al. analysed a GWAS previously conducted in asthmatic children7 and focused on the group with severe asthma. They found that certain variants of the PDCD4 gene (10q24) are significantly associated to severe childhood asthma and to total IgE levels. Gel retardation and reporter gene tests confirmed that the identified SNP alters binding of the transcription factor MYB, influencing the expression of PDCD4 – a known target of MYB.12
An association has recently been described between DNA methylation and asthma susceptibility dependent upon age and gender.13 The authors carried out a GWAS with the purpose of determining whether the association of the region of chromosome 17q12-q21 to childhood asthma was gender-specific. The study identified a regulatory region in the promoter of the ZPBP2 gene that showed statistically significant differences among males, females and age in terms of DNA methylation. The effect of the variants in 17q12-q21 upon asthma susceptibility could be attenuated by DNA methylation. Adult males and females carrying the 17q12-q21 alleles predisposing to asthma would not be affected, due to the high methylation levels of the ZPBP2 gene. The authors suggested that locus-specific DNA methylation could exert a modulating effect upon the development of asthma. The loci identified in the main GWAS published to date are summarised in Table 2.
The future of genetics in asthmaGenome-wide association studies have caused a genuine revolution in the study of complex diseases such as asthma. Over 1000 genes have been associated to asthma (Human Genetic Epidemiology Navigator Database, http://hugenavigator.net/HuGENavigator/home.do, search in September 2013), and may constitute potential therapeutic targets. However, despite such success, the results have fallen short of the expectations when it comes to explaining the genetic origin of the disease – a condition that has been referred to as “missing heritability”. This may be due to technological problems that preclude detection of all the SNPs or to copy number variations (CNV). Another hypothesis is that environmental factors interact with the genetic variants, influencing the development and severity of the disease. An example of this is the interaction between a polymorphism in the CD14 gene and exposure to a bacterial lipopolysaccharide, where the response of individuals with the same genetic burden depends on the polysaccharide levels in the environment.3 Genome wide interaction scans (GWIS) apply genetic variant searches taking into account environmental factors. A number of such studies have found new genes not detected in a first GWAS.2,3,14 On the other hand, GWAS in asthma have also been carried out taking into account gene–gene interactions, in which the variation of a locus alters the effect of the variation of a second locus reflecting epistasis between two or more genes. The gene–gene interaction studies are still in course, and it is not yet known whether this method will be able to explain the heritability of asthma. However, gene–environment interactions would in part help explain the mentioned “missing heritability”.2
Another issue that remains to be addressed is the association of genes of unknown function or which cannot be easily related to the disease, such as the ORMDL3/GSDM1L or DENND1B genes. The phenotypic heterogeneity of a disorder such as asthma could be decisive for establishing genuine genetic associations. Efforts to obtain more homogeneous asthma patient subgroups will serve to increase the performance of future GWAS in asthma.
In the coming years, technologies for the identification of genetic variants will continue to develop, giving rise to a tremendous increase in the volume of information. The combination of genome-wide studies with next generation sequencing (exome, whole genome, methylation) and re-sequencing techniques targeted to concrete regions of the genome will allow us to detect rare or infrequent variants that cannot be detected using microarray technology, and which contribute to the complexity of asthma. In fact, it is now believed that uncommon variants, not detected by GWAS, could make a greater contribution to risk – in contrast to the previously held idea that common diseases are associated to common genic variants. Most of the genic variants described are found in non-coding regions suggesting that gene regulation plays an important role in susceptibility to developing the disease. Sequencing the complete genome will help clarify this point.2,3
Epigenetic regulation is crucial in mediating environmental influences upon genic expression. We have recently described variations in the DNA methylation of a specific locus in the B cell population of a subgroup of asthmatic individuals.15 There have been descriptions of variations, including methylation, in the promoter of the PTGDR gene (Prostaglandin D2 Receptor) in asthmatics and atopic individuals.16 The combination of GWAS and promoter methylation, histone modifications and the influence upon the genic regulation on non-coding RNA is an extremely interesting approach that will open up new paths of research in asthma. The great amount of information produced by GWAS should serve to address functional genetics with a view to understanding the different pathways and genes that participate in the pathogenesis of asthma.
This work was supported by the Spanish “Fondo de Investigación Sanitaria” (FIS) grants (PI10/01706 and PI13/00564) that were cofounded with FEDER funds; by the Junta de Castilla y León (GRS/745/A/13 and BIO/SA67/13); “Fundación Botín-Universidad de Salamanca”; “Sociedad Española de Alergología e Inmunología Clínica”; and 2013 “Fundación Salud 2000” research allergology grants.